+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9770 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
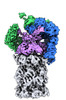
| Title | yeast proteasome in substrate-processing state (C3-b) | |||||||||
 Map data Map data | substrate-processing C3-b | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Proteasome / K48-Ub4 / Ub-bound / cryo-EM / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationSAGA complex localization to transcription regulatory region / : / proteasome regulatory particle assembly / proteasome storage granule assembly / transcription export complex 2 / peroxisome fission / protein deneddylation / maintenance of DNA trinucleotide repeats / filamentous growth / COP9 signalosome ...SAGA complex localization to transcription regulatory region / : / proteasome regulatory particle assembly / proteasome storage granule assembly / transcription export complex 2 / peroxisome fission / protein deneddylation / maintenance of DNA trinucleotide repeats / filamentous growth / COP9 signalosome / proteasome regulatory particle / protein-containing complex localization / mitochondrial fission / proteasome-activating activity / proteasome regulatory particle, lid subcomplex / proteasome regulatory particle, base subcomplex / metal-dependent deubiquitinase activity / ER-Phagosome pathway / Antigen processing: Ub, ATP-independent proteasomal degradation / K48-linked polyubiquitin modification-dependent protein binding / proteasome core complex assembly / nuclear outer membrane-endoplasmic reticulum membrane network / Proteasome assembly / Cross-presentation of soluble exogenous antigens (endosomes) / TNFR2 non-canonical NF-kB pathway / nonfunctional rRNA decay / Ubiquitin-Mediated Degradation of Phosphorylated Cdc25A / Regulation of PTEN stability and activity / proteasomal ubiquitin-independent protein catabolic process / CDK-mediated phosphorylation and removal of Cdc6 / FBXL7 down-regulates AURKA during mitotic entry and in early mitosis / KEAP1-NFE2L2 pathway / Neddylation / peptide catabolic process / proteasome binding / Orc1 removal from chromatin / MAPK6/MAPK4 signaling / regulation of protein catabolic process / proteasome storage granule / Antigen processing: Ubiquitination & Proteasome degradation / positive regulation of RNA polymerase II transcription preinitiation complex assembly / polyubiquitin modification-dependent protein binding / protein deubiquitination / proteasome endopeptidase complex / proteasome core complex, beta-subunit complex / Ub-specific processing proteases / endopeptidase activator activity / proteasome assembly / threonine-type endopeptidase activity / mRNA export from nucleus / proteasome core complex, alpha-subunit complex / enzyme regulator activity / ERAD pathway / Neutrophil degranulation / protein folding chaperone / proteasome complex / ubiquitin binding / nucleotide-excision repair / positive regulation of transcription elongation by RNA polymerase II / double-strand break repair via homologous recombination / metallopeptidase activity / positive regulation of protein catabolic process / peroxisome / ubiquitin-dependent protein catabolic process / endopeptidase activity / protein-macromolecule adaptor activity / molecular adaptor activity / proteasome-mediated ubiquitin-dependent protein catabolic process / ubiquitinyl hydrolase 1 / cysteine-type deubiquitinase activity / regulation of cell cycle / chromatin remodeling / protein domain specific binding / mRNA binding / ubiquitin protein ligase binding / endoplasmic reticulum membrane / structural molecule activity / endoplasmic reticulum / positive regulation of transcription by RNA polymerase II / ATP hydrolysis activity / mitochondrion / ATP binding / metal ion binding / identical protein binding / nucleus / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.5 Å | |||||||||
 Authors Authors | Cong Y | |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2019 Journal: Mol Cell / Year: 2019Title: Structural Snapshots of 26S Proteasome Reveal Tetraubiquitin-Induced Conformations. Authors: Zhanyu Ding / Cong Xu / Indrajit Sahu / Yifan Wang / Zhenglin Fu / Min Huang / Catherine C L Wong / Michael H Glickman / Yao Cong /   Abstract: The 26S proteasome is the ATP-dependent protease responsible for regulating the proteome of eukaryotic cells through degradation of mainly ubiquitin-tagged substrates. In order to understand how ...The 26S proteasome is the ATP-dependent protease responsible for regulating the proteome of eukaryotic cells through degradation of mainly ubiquitin-tagged substrates. In order to understand how proteasome responds to ubiquitin signal, we resolved an ensemble of cryo-EM structures of proteasome in the presence of K48-Ub, with three of them resolved at near-atomic resolution. We identified a conformation with stabilized ubiquitin receptors and a previously unreported orientation of the lid, assigned as a Ub-accepted state C1-b. We determined another structure C3-b with localized K48-Ub to the toroid region of Rpn1, assigned as a substrate-processing state. Our structures indicate that tetraUb induced conformational changes in proteasome could initiate substrate degradation. We also propose a CP gate-opening mechanism involving the propagation of the motion of the lid to the gate through the Rpn6-α2 interaction. Our results enabled us to put forward a model of a functional cycle for proteasomes induced by tetraUb and nucleotide. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9770.map.gz emd_9770.map.gz | 14.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9770-v30.xml emd-9770-v30.xml emd-9770.xml emd-9770.xml | 54 KB 54 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9770.png emd_9770.png | 149.9 KB | ||
| Filedesc metadata |  emd-9770.cif.gz emd-9770.cif.gz | 14.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9770 http://ftp.pdbj.org/pub/emdb/structures/EMD-9770 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9770 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9770 | HTTPS FTP |
-Related structure data
| Related structure data |  6j2nMC  9769C  9771C  9772C  9773C  6j2cC  6j2qC  6j2xC  6j30C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9770.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9770.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | substrate-processing C3-b | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.318 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
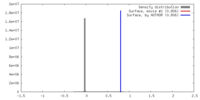
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : yeast 26S proteasome
+Supramolecule #1: yeast 26S proteasome
+Macromolecule #1: Proteasome subunit beta type-1
+Macromolecule #2: Proteasome subunit beta type-2
+Macromolecule #3: Proteasome subunit beta type-3
+Macromolecule #4: Proteasome subunit beta type-4
+Macromolecule #5: Proteasome subunit beta type-5
+Macromolecule #6: Proteasome subunit beta type-6
+Macromolecule #7: Proteasome subunit beta type-7
+Macromolecule #8: Proteasome subunit alpha type-1
+Macromolecule #9: Proteasome subunit alpha type-2
+Macromolecule #10: Proteasome subunit alpha type-3
+Macromolecule #11: Proteasome subunit alpha type-4
+Macromolecule #12: Proteasome subunit alpha type-5
+Macromolecule #13: Proteasome subunit alpha type-6
+Macromolecule #14: Probable proteasome subunit alpha type-7
+Macromolecule #15: 26S protease regulatory subunit 7 homolog
+Macromolecule #16: 26S protease regulatory subunit 4 homolog
+Macromolecule #17: 26S protease regulatory subunit 8 homolog
+Macromolecule #18: 26S protease regulatory subunit 6B homolog
+Macromolecule #19: 26S protease subunit RPT4
+Macromolecule #20: 26S protease regulatory subunit 6A
+Macromolecule #21: 26S proteasome regulatory subunit RPN2
+Macromolecule #22: 26S proteasome regulatory subunit RPN9
+Macromolecule #23: 26S proteasome regulatory subunit RPN5
+Macromolecule #24: 26S proteasome regulatory subunit RPN6
+Macromolecule #25: 26S proteasome regulatory subunit RPN7
+Macromolecule #26: 26S proteasome regulatory subunit RPN3
+Macromolecule #27: 26S proteasome regulatory subunit RPN12
+Macromolecule #28: 26S proteasome regulatory subunit RPN8
+Macromolecule #29: Ubiquitin carboxyl-terminal hydrolase RPN11
+Macromolecule #30: 26S proteasome regulatory subunit RPN10
+Macromolecule #31: 26S proteasome regulatory subunit RPN13
+Macromolecule #32: 26S proteasome complex subunit SEM1
+Macromolecule #33: 26S proteasome regulatory subunit RPN1
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 38.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)