-Search query
-Search result
Showing 1 - 50 of 62 items for (author: wenli & z)
EMDB-35730: 
Cryo-EM structure of SARS-CoV-2 spike protein in complex with 8H12

EMDB-35731: 
Cryo-EM structure of SARS-CoV-2 spike protein in complex with 3E2

EMDB-35736: 
Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 1C4
EMDB-35739: 
Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 3E2 and 1C4

EMDB-35740: 
Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 3E2 (local refinement)

EMDB-35741: 
Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 1C4 (local refinement)

EMDB-35743: 
Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 3E2
EMDB-35745: 
cryo-EM structure of SARS-CoV-2 Omicron BA.1 spike protein in complex with double nAbs 8H12 and 3E2
EMDB-35746: 
Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 3E2 and 1C4 (local refinement)

EMDB-35747: 
cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with double nAbs 8H12 and 3E2
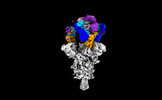
EMDB-35749: 
Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs XMA01 and 3E2

EMDB-35750: 
cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs S2E12 and 3E2

EMDB-35751: 
Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs S2E12 and 3E2 (local refinement)

EMDB-35752: 
cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and S2X35
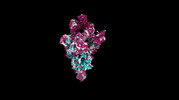
EMDB-35753: 
cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and VacW-209
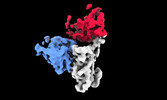
EMDB-35754: 
cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and S2X35 (local refinement)
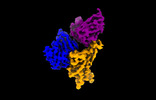
EMDB-35755: 
Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs XMA01 and 3E2 (local refinement)

PDB-8iv4: 
Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 3E2 (local refinement)

PDB-8iv5: 
Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 1C4 (local refinement)

PDB-8iv8: 
Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 3E2 and 1C4 (local refinement)

PDB-8iva: 
Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs XMA01 and 3E2 (local refinement)

EMDB-27920: 
3H03 Fab in complex with influenza virus neuraminidase from A/Brevig Mission/1/1918 (H1N1)

EMDB-27921: 
2H08 Fab in complex with influenza virus neuraminidase from A/Brevig Mission/1/1918 (H1N1)

PDB-8e6j: 
3H03 Fab in complex with influenza virus neuraminidase from A/Brevig Mission/1/1918 (H1N1)

PDB-8e6k: 
2H08 Fab in complex with influenza virus neuraminidase from A/Brevig Mission/1/1918 (H1N1)

EMDB-33247: 
Cryo-EM structure of the Neuromedin U receptor 2 (NMUR2) in complex with G Protein and its endogeneous Peptide-Agonist NMU25

PDB-7xk8: 
Cryo-EM structure of the Neuromedin U receptor 2 (NMUR2) in complex with G Protein and its endogeneous Peptide-Agonist NMU25

EMDB-33302: 
CryoEM structure of the somatostatin receptor 2 (SSTR2) in complex with Gi1 and its endogeneous peptide ligand SST-14

EMDB-33303: 
CryoEM structure of somatostatin receptor 4 (SSTR4) in complex with Gi1 and its endogeneous ligand SST-14

EMDB-33304: 
CryoEM structure of somatostatin receptor 4 (SSTR4) with Gi1 and J-2156

PDB-7xmr: 
CryoEM structure of the somatostatin receptor 2 (SSTR2) in complex with Gi1 and its endogeneous peptide ligand SST-14

PDB-7xms: 
CryoEM structure of somatostatin receptor 4 (SSTR4) in complex with Gi1 and its endogeneous ligand SST-14

PDB-7xmt: 
CryoEM structure of somatostatin receptor 4 (SSTR4) with Gi1 and J-2156

EMDB-33107: 
Cryo-EM structure of the human chemokine receptor CX3CR1 in complex with Gi1

EMDB-33108: 
Cryo-EM structure of the human chemokine receptor CX3CR1 in complex with CX3CL1 and Gi1

PDB-7xbw: 
Cryo-EM structure of the human chemokine receptor CX3CR1 in complex with Gi1

PDB-7xbx: 
Cryo-EM structure of the human chemokine receptor CX3CR1 in complex with CX3CL1 and Gi1

EMDB-26928: 
Negative stain map of cH4/3
EMDB-26929: 
Negative stain half map of cH4/3 state 1

EMDB-26930: 
Negative stain map of cH4/3 state 3

EMDB-26931: 
Negative stain map of cH4/3 state 3

EMDB-26932: 
Negative stain map of cH4/3 state 4
EMDB-26933: 
Negative stain map of cH15/3

EMDB-26934: 
Negative stain map of cH15/3 state 1
EMDB-26935: 
Negative stain map of cH15/3 state 2

EMDB-26937: 
Negative stain map of cH15/3 state 3

EMDB-26938: 
Negative stain map of cH15/3 state 4
EMDB-26939: 
Negative stain map of cH15/3 in complex with CR9114 Fab

EMDB-31524: 
Structure of the SARS-CoV-2 A372T spike glycoprotein (open)

EMDB-31525: 
Structure of the SARS-CoV-2 A372T spike glycoprotein (closed)
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
