[English] 日本語
 Yorodumi
Yorodumi- EMDB-8640: Asymmetric structure of AcrAB-TolC tripartite multi drug efflux pump -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8640 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Asymmetric structure of AcrAB-TolC tripartite multi drug efflux pump | |||||||||
 Map data Map data | AcrABZ-TolC with puromycin | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationMacAB-TolC complex / enterobactin transport / bile acid transmembrane transporter activity / xenobiotic detoxification by transmembrane export across the cell outer membrane /  efflux pump complex / periplasmic side of plasma membrane / xenobiotic detoxification by transmembrane export across the plasma membrane / bile acid and bile salt transport / efflux pump complex / periplasmic side of plasma membrane / xenobiotic detoxification by transmembrane export across the plasma membrane / bile acid and bile salt transport /  porin activity / xenobiotic transmembrane transporter activity ...MacAB-TolC complex / enterobactin transport / bile acid transmembrane transporter activity / xenobiotic detoxification by transmembrane export across the cell outer membrane / porin activity / xenobiotic transmembrane transporter activity ...MacAB-TolC complex / enterobactin transport / bile acid transmembrane transporter activity / xenobiotic detoxification by transmembrane export across the cell outer membrane /  efflux pump complex / periplasmic side of plasma membrane / xenobiotic detoxification by transmembrane export across the plasma membrane / bile acid and bile salt transport / efflux pump complex / periplasmic side of plasma membrane / xenobiotic detoxification by transmembrane export across the plasma membrane / bile acid and bile salt transport /  porin activity / xenobiotic transmembrane transporter activity / efflux transmembrane transporter activity / transmembrane transporter activity / monoatomic ion transmembrane transport / cell outer membrane / response to organic cyclic compound / response to toxic substance / outer membrane-bounded periplasmic space / monoatomic ion channel activity / response to xenobiotic stimulus / response to antibiotic / porin activity / xenobiotic transmembrane transporter activity / efflux transmembrane transporter activity / transmembrane transporter activity / monoatomic ion transmembrane transport / cell outer membrane / response to organic cyclic compound / response to toxic substance / outer membrane-bounded periplasmic space / monoatomic ion channel activity / response to xenobiotic stimulus / response to antibiotic /  membrane / identical protein binding / membrane / identical protein binding /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Escherichia coli K12 (bacteria) / Escherichia coli K12 (bacteria) /   Escherichia coli (strain K12) (bacteria) Escherichia coli (strain K12) (bacteria) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 5.9 Å cryo EM / Resolution: 5.9 Å | |||||||||
 Authors Authors | Wang Z / Fan G / Hryc CF / Blaza JN / Serysheva II / Schmid MF / Chiu W / Luisi BF / Du D | |||||||||
 Citation Citation |  Journal: Elife / Year: 2017 Journal: Elife / Year: 2017Title: An allosteric transport mechanism for the AcrAB-TolC multidrug efflux pump. Authors: Zhao Wang / Guizhen Fan / Corey F Hryc / James N Blaza / Irina I Serysheva / Michael F Schmid / Wah Chiu / Ben F Luisi / Dijun Du /   Abstract: Bacterial efflux pumps confer multidrug resistance by transporting diverse antibiotics from the cell. In Gram-negative bacteria, some of these pumps form multi-protein assemblies that span the cell ...Bacterial efflux pumps confer multidrug resistance by transporting diverse antibiotics from the cell. In Gram-negative bacteria, some of these pumps form multi-protein assemblies that span the cell envelope. Here, we report the near-atomic resolution cryoEM structures of the AcrAB-TolC multidrug efflux pump in resting and drug transport states, revealing a quaternary structural switch that allosterically couples and synchronizes initial ligand binding with channel opening. Within the transport-activated state, the channel remains open even though the pump cycles through three distinct conformations. Collectively, our data provide a dynamic mechanism for the assembly and operation of the AcrAB-TolC pump. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8640.map.gz emd_8640.map.gz | 59.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8640-v30.xml emd-8640-v30.xml emd-8640.xml emd-8640.xml | 14.4 KB 14.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8640_fsc.xml emd_8640_fsc.xml | 8.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_8640.png emd_8640.png | 76.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8640 http://ftp.pdbj.org/pub/emdb/structures/EMD-8640 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8640 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8640 | HTTPS FTP |
-Related structure data
| Related structure data |  5o66MC  3636C  8636C  5nc5C  5ng5C  5v5sC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8640.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8640.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | AcrABZ-TolC with puromycin | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.62 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : AcrABZ-TolC
| Entire | Name: AcrABZ-TolC |
|---|---|
| Components |
|
-Supramolecule #1: AcrABZ-TolC
| Supramolecule | Name: AcrABZ-TolC / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:   Escherichia coli K12 (bacteria) Escherichia coli K12 (bacteria) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Macromolecule #1: Multidrug efflux pump subunit AcrA
| Macromolecule | Name: Multidrug efflux pump subunit AcrA / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli (strain K12) (bacteria) Escherichia coli (strain K12) (bacteria) |
| Molecular weight | Theoretical: 39.80066 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: CDDKQAQQGG QQMPAVGVVT VKTEPLQITT ELPGRTSAYR IAEVRPQVSG IILKRNFKEG SDIEAGVSLY QIDPATYQAT YDSAKGDLA KAQAAANIAQ LTVNRYQKLL GTQYISKQEY DQALADAQQA NAAVTAAKAA VETARINLAY TKVTSPISGR I GKSNVTEG ...String: CDDKQAQQGG QQMPAVGVVT VKTEPLQITT ELPGRTSAYR IAEVRPQVSG IILKRNFKEG SDIEAGVSLY QIDPATYQAT YDSAKGDLA KAQAAANIAQ LTVNRYQKLL GTQYISKQEY DQALADAQQA NAAVTAAKAA VETARINLAY TKVTSPISGR I GKSNVTEG ALVQNGQATA LATVQQLDPI YVDVTQSSND MMRLKQELAN GTLKQENGKA KVSLITSDGI KFPQDGTLEF SD VTVDQTT GSITLRAIFP NPDHTMMPGM FVRARLEEGL NPNAILVPQQ GVTRTPRGDA TVLVVGADDK VETRPIVASQ AIG DKWLVT EGLKAGDRVV ISGLQKVRPG VQVKAQEVTA DNNQQAASGA QPEQSKS |
-Macromolecule #2: Outer membrane protein TolC
| Macromolecule | Name: Outer membrane protein TolC / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli (strain K12) (bacteria) / Strain: K12 Escherichia coli (strain K12) (bacteria) / Strain: K12 |
| Molecular weight | Theoretical: 53.783355 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MKKLLPILIG LSLSGFSSLS QAENLMQVYQ QARLSNPELR KSAADRDAAF EKINEARSPL LPQLGLGADY TYSNGYRDAN GINSNATSA SLQLTQSIFD MSKWRALTLQ EKAAGIQDVT YQTDQQTLIL NTATAYFNVL NAIDVLSYTQ AQKEAIYRQL D QTTQRFNV ...String: MKKLLPILIG LSLSGFSSLS QAENLMQVYQ QARLSNPELR KSAADRDAAF EKINEARSPL LPQLGLGADY TYSNGYRDAN GINSNATSA SLQLTQSIFD MSKWRALTLQ EKAAGIQDVT YQTDQQTLIL NTATAYFNVL NAIDVLSYTQ AQKEAIYRQL D QTTQRFNV GLVAITDVQN ARAQYDTVLA NEVTARNNLD NAVEQLRQIT GNYYPELAAL NVENFKTDKP QPVNALLKEA EK RNLSLLQ ARLSQDLARE QIRQAQDGHL PTLDLTASTG ISDTSYSGSK TRGAAGTQYD DSNMGQNKVG LSFSLPIYQG GMV NSQVKQ AQYNFVGASE QLESAHRSVV QTVRSSFNNI NASISSINAY KQAVVSAQSS LDAMEAGYSV GTRTIVDVLD ATTT LYNAK QELANARYNY LINQLNIKSA LGTLNEQDLL ALNNALSKPV STNPENVAPQ TPEQNAIADG YAPDSPAPVV QQTSA RTTT SNGHNPFRN |
-Macromolecule #3: Multidrug efflux pump subunit AcrB
| Macromolecule | Name: Multidrug efflux pump subunit AcrB / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli (strain K12) (bacteria) / Strain: K12 Escherichia coli (strain K12) (bacteria) / Strain: K12 |
| Molecular weight | Theoretical: 113.66518 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MPNFFIDRPI FAWVIAIIIM LAGGLAILKL PVAQYPTIAP PAVTISASYP GADAKTVQDT VTQVIEQNMN GIDNLMYMSS NSDSTGTVQ ITLTFESGTD ADIAQVQVQN KLQLAMPLLP QEVQQQGVSV EKSSSSFLMV VGVINTDGTM TQEDISDYVA A NMKDAISR ...String: MPNFFIDRPI FAWVIAIIIM LAGGLAILKL PVAQYPTIAP PAVTISASYP GADAKTVQDT VTQVIEQNMN GIDNLMYMSS NSDSTGTVQ ITLTFESGTD ADIAQVQVQN KLQLAMPLLP QEVQQQGVSV EKSSSSFLMV VGVINTDGTM TQEDISDYVA A NMKDAISR TSGVGDVQLF GSQYAMRIWM NPNELNKFQL TPVDVITAIK AQNAQVAAGQ LGGTPPVKGQ QLNASIIAQT RL TSTEEFG KILLKVNQDG SRVLLRDVAK IELGGENYDI IAEFNGQPAS GLGIKLATGA NALDTAAAIR AELAKMEPFF PSG LKIVYP YDTTPFVKIS IHEVVKTLVE AIILVFLVMY LFLQNFRATL IPTIAVPVVL LGTFAVLAAF GFSINTLTMF GMVL AIGLL VDDAIVVVEN VERVMAEEGL PPKEATRKSM GQIQGALVGI AMVLSAVFVP MAFFGGSTGA IYRQFSITIV SAMAL SVLV ALILTPALCA TMLKPIAKGD HGEGKKGFFG WFNRMFEKST HHYTDSVGGI LRSTGRYLVL YLIIVVGMAY LFVRLP SSF LPDEDQGVFM TMVQLPAGAT QERTQKVLNE VTHYYLTKEK NNVESVFAVN GFGFAGRGQN TGIAFVSLKD WADRPGE EN KVEAITMRAT RAFSQIKDAM VFAFNLPAIV ELGTATGFDF ELIDQAGLGH EKLTQARNQL LAEAAKHPDM LTSVRPNG L EDTPQFKIDI DQEKAQALGV SINDINTTLG AAWGGSYVND FIDRGRVKKV YVMSEAKYRM LPDDIGDWYV RAADGQMVP FSAFSSSRWE YGSPRLERYN GLPSMEILGQ AAPGKSTGEA MELMEQLASK LPTGVGYDWT GMSYQERLSG NQAPSLYAIS LIVVFLCLA ALYESWSIPF SVMLVVPLGV IGALLAATFR GLTNDVYFQV GLLTTIGLSA KNAILIVEFA KDLMDKEGKG L IEATLDAV RMRLRPILMT SLAFILGVMP LVISTGAGSG AQNAVGTGVM GGMVTATVLA IFFVPVFFVV VRRRFSRKNE DI EHSHTVD HH |
-Macromolecule #4: Multidrug efflux pump accessory protein AcrZ
| Macromolecule | Name: Multidrug efflux pump accessory protein AcrZ / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli (strain K12) (bacteria) Escherichia coli (strain K12) (bacteria) |
| Molecular weight | Theoretical: 5.995157 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MLELLKSLVF AVIMVPVVMA IILGLIYGLG EVFNIFSGVG KKDQPGQNHH HHHH |
-Macromolecule #5: 6-[2-(3,4-dimethoxyphenyl)ethylsulfanyl]-8-[4-(2-methoxyethyl)pip...
| Macromolecule | Name: 6-[2-(3,4-dimethoxyphenyl)ethylsulfanyl]-8-[4-(2-methoxyethyl)piperazin-1-yl]-3,3-dimethyl-1,4-dihydropyrano[3,4-c]pyridine-5-carbonitrile type: ligand / ID: 5 / Number of copies: 3 / Formula: 5QF |
|---|---|
| Molecular weight | Theoretical: 526.691 Da |
| Chemical component information | 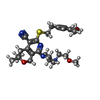 ChemComp-5QF: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 22.0 e/Å2 |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller