+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7030 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Staphylococcus aureus phage 80alpha procapsid | |||||||||
 Map data Map data | Capsid protein and C-terminal part of scaffolding protein in the Staphylococcus aureus phage 80alpha procapsid: clipped map used for model building and refinement in Coot and REFMAC | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | major capsid protein / HK97-like fold /  scaffolding protein / scaffolding protein /  procapsid / procapsid /  VIRUS VIRUS | |||||||||
| Function / homology | Protein of unknown function DUF4355 / Domain of unknown function (DUF4355) / viral scaffold / Phage capsid / Phage capsid family /  Scaffold protein / Major capsid protein Scaffold protein / Major capsid protein Function and homology information Function and homology information | |||||||||
| Biological species |  Staphylococcus phage 80alpha (virus) Staphylococcus phage 80alpha (virus) | |||||||||
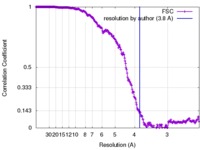
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.8 Å cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Kizziah JL / Dearborn AD | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2017 Journal: Elife / Year: 2017Title: Competing scaffolding proteins determine capsid size during mobilization of pathogenicity islands. Authors: Altaira D Dearborn / Erin A Wall / James L Kizziah / Laura Klenow / Laura K Parker / Keith A Manning / Michael S Spilman / John M Spear / Gail E Christie / Terje Dokland /  Abstract: pathogenicity islands (SaPIs), such as SaPI1, exploit specific helper bacteriophages, like 80α, for their high frequency mobilization, a process termed 'molecular piracy'. SaPI1 redirects the ... pathogenicity islands (SaPIs), such as SaPI1, exploit specific helper bacteriophages, like 80α, for their high frequency mobilization, a process termed 'molecular piracy'. SaPI1 redirects the helper's assembly pathway to form small capsids that can only accommodate the smaller SaPI1 genome, but not a complete phage genome. SaPI1 encodes two proteins, CpmA and CpmB, that are responsible for this size redirection. We have determined the structures of the 80α and SaPI1 procapsids to near-atomic resolution by cryo-electron microscopy, and show that CpmB competes with the 80α scaffolding protein (SP) for a binding site on the capsid protein (CP), and works by altering the angle between capsomers. We probed these interactions genetically and identified second-site suppressors of lethal mutations in SP. Our structures show, for the first time, the detailed interactions between SP and CP in a bacteriophage, providing unique insights into macromolecular assembly processes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7030.map.gz emd_7030.map.gz | 199.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7030-v30.xml emd-7030-v30.xml emd-7030.xml emd-7030.xml | 14.1 KB 14.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7030_fsc.xml emd_7030_fsc.xml | 26.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_7030.png emd_7030.png | 300.2 KB | ||
| Filedesc metadata |  emd-7030.cif.gz emd-7030.cif.gz | 5.6 KB | ||
| Others |  emd_7030_additional.map.gz emd_7030_additional.map.gz | 974.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7030 http://ftp.pdbj.org/pub/emdb/structures/EMD-7030 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7030 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7030 | HTTPS FTP |
-Related structure data
| Related structure data |  6b0xMC  7035C  6b23C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7030.map.gz / Format: CCP4 / Size: 210.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7030.map.gz / Format: CCP4 / Size: 210.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Capsid protein and C-terminal part of scaffolding protein in the Staphylococcus aureus phage 80alpha procapsid: clipped map used for model building and refinement in Coot and REFMAC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.21 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Capsid protein and C-terminal part of scaffolding protein...
| File | emd_7030_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Capsid protein and C-terminal part of scaffolding protein in the Staphylococcus aureus phage 80alpha procapsid: final map sharpened with a B-factor of 300 to 6 Angstrom resolution (cutoff at 3 Angstrom) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Staphylococcus phage 80alpha
| Entire | Name:  Staphylococcus phage 80alpha (virus) Staphylococcus phage 80alpha (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Staphylococcus phage 80alpha
| Supramolecule | Name: Staphylococcus phage 80alpha / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: Lysogenic 80alpha with small terminase gene deletion NCBI-ID: 53369 / Sci species name: Staphylococcus phage 80alpha / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:   Staphylococcus aureus (bacteria) Staphylococcus aureus (bacteria) |
| Molecular weight | Theoretical: 25.27 MDa |
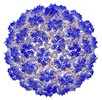
| Virus shell | Shell ID: 1 / Name: Procapsid / Diameter: 546.0 Å / T number (triangulation number): 7 |
-Macromolecule #1: Major head protein
| Macromolecule | Name: Major head protein / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Staphylococcus phage 80alpha (virus) Staphylococcus phage 80alpha (virus) |
| Molecular weight | Theoretical: 36.846883 KDa |
| Recombinant expression | Organism:   Staphylococcus aureus (bacteria) Staphylococcus aureus (bacteria) |
| Sequence | String: MEQTQKLKLN LQHFASNNVK PQVFNPDNVM MHEKKDGTLM NEFTTPILQE VMENSKIMQL GKYEPMEGTE KKFTFWADKP GAYWVGEGQ KIETSKATWV NATMRAFKLG VILPVTKEFL NYTYSQFFEE MKPMIAEAFY KKFDEAGILN QGNNPFGKSI A QSIEKTNK ...String: MEQTQKLKLN LQHFASNNVK PQVFNPDNVM MHEKKDGTLM NEFTTPILQE VMENSKIMQL GKYEPMEGTE KKFTFWADKP GAYWVGEGQ KIETSKATWV NATMRAFKLG VILPVTKEFL NYTYSQFFEE MKPMIAEAFY KKFDEAGILN QGNNPFGKSI A QSIEKTNK VIKGDFTQDN IIDLEALLED DELEANAFIS KTQNRSLLRK IVDPETKERI YDRNSDSLDG LPVVNLKSSN LK RGELITG DFDKLIYGIP QLIEYKIDET AQLSTVKNED GTPVNLFEQD MVALRATMHV ALHIADDKAF AKLVPADKRT DSV PGEV UniProtKB: Major capsid protein |
-Macromolecule #2: Scaffold protein
| Macromolecule | Name: Scaffold protein / type: protein_or_peptide / ID: 2 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Staphylococcus phage 80alpha (virus) Staphylococcus phage 80alpha (virus) |
| Molecular weight | Theoretical: 23.410941 KDa |
| Recombinant expression | Organism:   Staphylococcus aureus (bacteria) Staphylococcus aureus (bacteria) |
| Sequence | String: MEENKLKFNL QFFADQSDDP DEPGGDGKKG NPDKKENDEG TEITFTPEQQ KKVDEILERR VAHEKKKADE YAKEKAAEAA KEAAKLAKM NKDQKDEYER EQMEKELEQL RSEKQLNEMR SEARKMLSEA EVDSSDEVVN LVVTDTAEQT KSNVEAFSNA V KKAVNEAV ...String: MEENKLKFNL QFFADQSDDP DEPGGDGKKG NPDKKENDEG TEITFTPEQQ KKVDEILERR VAHEKKKADE YAKEKAAEAA KEAAKLAKM NKDQKDEYER EQMEKELEQL RSEKQLNEMR SEARKMLSEA EVDSSDEVVN LVVTDTAEQT KSNVEAFSNA V KKAVNEAV KVNARQSPLT GGDSFNHSTK NKPQNLAEIA RQKRIIKN UniProtKB:  Scaffold protein Scaffold protein |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Image recording | Film or detector model: DIRECT ELECTRON DE-20 (5k x 3k) / Average electron dose: 40.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z
Z Y
Y X
X