+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9877 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of PSI dimer from Anabaena | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Nostoc sp. PCC 7120 (bacteria) Nostoc sp. PCC 7120 (bacteria) | |||||||||
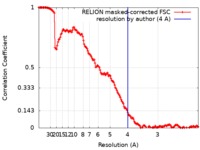
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Kato K / Nagao R / Shen JR / Miyazaki N / Akita F | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: Structure of a cyanobacterial photosystem I tetramer revealed by cryo-electron microscopy. Authors: Koji Kato / Ryo Nagao / Tian-Yi Jiang / Yoshifumi Ueno / Makio Yokono / Siu Kit Chan / Mai Watanabe / Masahiko Ikeuchi / Jian-Ren Shen / Seiji Akimoto / Naoyuki Miyazaki / Fusamichi Akita /  Abstract: Photosystem I (PSI) functions to harvest light energy for conversion into chemical energy. The organisation of PSI is variable depending on the species of organism. Here we report the structure of a ...Photosystem I (PSI) functions to harvest light energy for conversion into chemical energy. The organisation of PSI is variable depending on the species of organism. Here we report the structure of a tetrameric PSI core isolated from a cyanobacterium, Anabaena sp. PCC 7120, analysed by single-particle cryo-electron microscopy (cryo-EM) at 3.3 Å resolution. The PSI tetramer has a C2 symmetry and is organised in a dimer of dimers form. The structure reveals interactions at the dimer-dimer interface and the existence of characteristic pigment orientations and inter-pigment distances within the dimer units that are important for unique excitation energy transfer. In particular, characteristic residues of PsaL are identified to be responsible for the formation of the tetramer. Time-resolved fluorescence analyses showed that the PSI tetramer has an enhanced excitation-energy quenching. These structural and spectroscopic findings provide insights into the physiological significance of the PSI tetramer and evolutionary changes of the PSI organisations. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9877.map.gz emd_9877.map.gz | 19.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9877-v30.xml emd-9877-v30.xml emd-9877.xml emd-9877.xml | 12.2 KB 12.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9877_fsc.xml emd_9877_fsc.xml | 15.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_9877.png emd_9877.png | 61.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9877 http://ftp.pdbj.org/pub/emdb/structures/EMD-9877 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9877 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9877 | HTTPS FTP |
-Validation report
| Summary document |  emd_9877_validation.pdf.gz emd_9877_validation.pdf.gz | 77.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9877_full_validation.pdf.gz emd_9877_full_validation.pdf.gz | 76.7 KB | Display | |
| Data in XML |  emd_9877_validation.xml.gz emd_9877_validation.xml.gz | 493 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9877 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9877 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9877 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9877 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9877.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9877.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.12 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : PSI dimer
| Entire | Name: PSI dimer |
|---|---|
| Components |
|
-Supramolecule #1: PSI dimer
| Supramolecule | Name: PSI dimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#12 |
|---|---|
| Source (natural) | Organism:  Nostoc sp. PCC 7120 (bacteria) Nostoc sp. PCC 7120 (bacteria) |
| Molecular weight | Theoretical: 700 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.014 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 6.5 / Component:
| ||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: Correlation coefficient |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)