+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9663 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
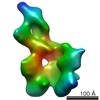
| Title | Saccharomyces cerevisiae SAGA complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationSLIK (SAGA-like) complex / SAGA complex / NuA4 histone acetyltransferase complex / Ub-specific processing proteases / chromatin remodeling / DNA repair / DNA-templated transcription / regulation of DNA-templated transcription / regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II / nucleus Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.9 Å | |||||||||
 Authors Authors | Zheng XD / Liu GC / Guan HP / Li HT | |||||||||
 Citation Citation |  Journal: Cell Discov / Year: 2019 Journal: Cell Discov / Year: 2019Title: Architecture of SAGA complex. Authors: Gaochao Liu / Xiangdong Zheng / Haipeng Guan / Yong Cao / Hongyuan Qu / Junqing Kang / Xiangle Ren / Jianlin Lei / Meng-Qiu Dong / Xueming Li / Haitao Li /  | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9663.map.gz emd_9663.map.gz | 10 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9663-v30.xml emd-9663-v30.xml emd-9663.xml emd-9663.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9663_fsc.xml emd_9663_fsc.xml | 12.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_9663.png emd_9663.png | 38.5 KB | ||
| Masks |  emd_9663_msk_1.map emd_9663_msk_1.map | 178 MB |  Mask map Mask map | |
| Others |  emd_9663_additional.map.gz emd_9663_additional.map.gz emd_9663_half_map_1.map.gz emd_9663_half_map_1.map.gz emd_9663_half_map_2.map.gz emd_9663_half_map_2.map.gz | 10 MB 141 MB 141 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9663 http://ftp.pdbj.org/pub/emdb/structures/EMD-9663 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9663 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9663 | HTTPS FTP |
-Validation report
| Summary document |  emd_9663_validation.pdf.gz emd_9663_validation.pdf.gz | 78.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9663_full_validation.pdf.gz emd_9663_full_validation.pdf.gz | 77.8 KB | Display | |
| Data in XML |  emd_9663_validation.xml.gz emd_9663_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9663 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9663 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9663 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9663 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9663.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9663.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.401 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_9663_msk_1.map emd_9663_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
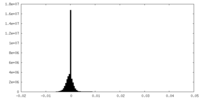
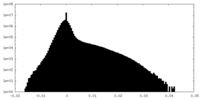
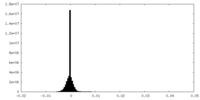
| Density Histograms |
-Additional map: Z-plane reversed map, for docking and comparison.
| File | emd_9663_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Z-plane reversed map, for docking and comparison. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_9663_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_9663_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Saccharomyces cerevisiae SAGA (Spt-Ada-Gcn5-Acetyltransferase) complex
| Entire | Name: Saccharomyces cerevisiae SAGA (Spt-Ada-Gcn5-Acetyltransferase) complex |
|---|---|
| Components |
|
-Supramolecule #1: Saccharomyces cerevisiae SAGA (Spt-Ada-Gcn5-Acetyltransferase) complex
| Supramolecule | Name: Saccharomyces cerevisiae SAGA (Spt-Ada-Gcn5-Acetyltransferase) complex type: complex / ID: 1 / Parent: 0 / Details: The multi-functional co-activator SAGA complex |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1.8 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 8.5 / Component:
| ||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 5.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 101.325 kPa Details: The grid was coated with a home-made thin continuous carbon film. | ||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 70.0 K / Max: 70.0 K |
| Specialist optics | Spherical aberration corrector: Cs-corrector are used |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Sampling interval: 5.0 µm / Digitization - Frames/image: 3-30 / Number real images: 8526 / Average exposure time: 0.25 sec. / Average electron dose: 5.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 3.0 µm / Calibrated defocus min: 1.2 µm / Calibrated magnification: 81000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.1 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)