[English] 日本語
 Yorodumi
Yorodumi- EMDB-9267: Single-Molecule 3D Image of DNA Origami Bennett Linkage by Image ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9267 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single-Molecule 3D Image of DNA Origami Bennett Linkage by Image Contrast Enhancement and Individual Particle Electron Tomography (No. 02) | |||||||||
 Map data Map data | Origami Bennett Linkage (No. 02) | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Enterobacteria phage M13 (virus) / synthetic construct (others) Enterobacteria phage M13 (virus) / synthetic construct (others) | |||||||||
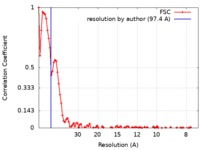
| Method | electron tomography / cryo EM / negative staining / Resolution: 97.4 Å | |||||||||
 Authors Authors | Wu H / Zhai X / Lei D / Liu J / Yu Y / Bie R / Ren G | |||||||||
 Citation Citation |  Journal: Sci Rep / Year: 2018 Journal: Sci Rep / Year: 2018Title: An Algorithm for Enhancing the Image Contrast of Electron Tomography. Authors: Hao Wu / Xiaobo Zhai / Dongsheng Lei / Jianfang Liu / Yadong Yu / Rongfang Bie / Gang Ren /   Abstract: Three-dimensional (3D) reconstruction of a single protein molecule is essential for understanding the relationship between the structural dynamics and functions of the protein. Electron tomography ...Three-dimensional (3D) reconstruction of a single protein molecule is essential for understanding the relationship between the structural dynamics and functions of the protein. Electron tomography (ET) provides a tool for imaging an individual particle of protein from a series of tilted angles. Individual-particle electron tomography (IPET) provides an approach for reconstructing a 3D density map from a single targeted protein particle (without averaging from different particles of this type of protein), in which the target particle was imaged from a series of tilting angles. However, owing to radiation damage limitations, low-dose images (high noise, and low image contrast) are often challenging to be aligned for 3D reconstruction at intermediate resolution (1-3 nm). Here, we propose a computational method to enhance the image contrast, without increasing any experimental dose, for IPET 3D reconstruction. Using an edge-preserving smoothing-based multi-scale image decomposition algorithm, this method can detect the object against a high-noise background and enhance the object image contrast without increasing the noise level or significantly decreasing the image resolution. The method was validated by using both negative staining (NS) ET and cryo-ET images. The successful 3D reconstruction of a small molecule (<100 kDa) indicated that this method can be used as a supporting tool to current ET 3D reconstruction methods for studying protein dynamics via structure determination from each individual particle of the same type of protein. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9267.map.gz emd_9267.map.gz | 59.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9267-v30.xml emd-9267-v30.xml emd-9267.xml emd-9267.xml | 10.1 KB 10.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9267_fsc.xml emd_9267_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_9267.png emd_9267.png | 17.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9267 http://ftp.pdbj.org/pub/emdb/structures/EMD-9267 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9267 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9267 | HTTPS FTP |
-Validation report
| Summary document |  emd_9267_validation.pdf.gz emd_9267_validation.pdf.gz | 79.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9267_full_validation.pdf.gz emd_9267_full_validation.pdf.gz | 78.7 KB | Display | |
| Data in XML |  emd_9267_validation.xml.gz emd_9267_validation.xml.gz | 496 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9267 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9267 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9267 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9267 | HTTPS FTP |
-Related structure data
| Related structure data |  9262C  9263C  9266C  9268C  9269C  9270C  9271C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9267.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9267.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Origami Bennett Linkage (No. 02) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.7 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : DNA origami Bennett linkage
| Entire | Name: DNA origami Bennett linkage |
|---|---|
| Components |
|
-Supramolecule #1: DNA origami Bennett linkage
| Supramolecule | Name: DNA origami Bennett linkage / type: complex / ID: 1 / Parent: 0 |
|---|
-Supramolecule #2: M13 phage genome segment
| Supramolecule | Name: M13 phage genome segment / type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage M13 (virus) Enterobacteria phage M13 (virus) |
| Recombinant expression | Organism:  |
-Supramolecule #3: Synthetic DNA oligonucleotides
| Supramolecule | Name: Synthetic DNA oligonucleotides / type: complex / ID: 3 / Parent: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Details: Tris-Borate-EDTA (TBE) buffer containing 11 mM MgCl2 |
|---|---|
| Staining | Type: POSITIVE / Material: Uranyl Formate Details: The grid was washed twice by 1% (w/v) uranyl formate |
| Grid | Model: Homemade / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: LACEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Instrument: LEICA EM GP |
| Details | DNA origami Bennett linkage was diluted to ~2 nM with Tris-Borate-EDTA (TBE) buffer containing 11 mM MgCl2 |
| Sectioning | Other: NO SECTIONING |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 0.69 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal magnification: 19000 |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)