+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9211 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Rabbit muscle aldolase using curated picks | |||||||||
 Map data Map data | Cryosparc v0 with curated picks sharpened | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.63 Å | |||||||||
 Authors Authors | Bepler T / Morin A / Brasch J / Shapiro L / Noble AJ / Berger B | |||||||||
 Citation Citation |  Journal: Res Comput Mol Biol / Year: 2018 Journal: Res Comput Mol Biol / Year: 2018Title: Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Authors: Tristan Bepler / Andrew Morin / Alex J Noble / Julia Brasch / Lawrence Shapiro / Bonnie Berger /  | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9211.map.gz emd_9211.map.gz | 59.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9211-v30.xml emd-9211-v30.xml emd-9211.xml emd-9211.xml | 17.4 KB 17.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9211.png emd_9211.png | 119.5 KB | ||
| Masks |  emd_9211_msk_1.map emd_9211_msk_1.map | 64 MB |  Mask map Mask map | |
| Others |  emd_9211_additional.map.gz emd_9211_additional.map.gz emd_9211_half_map_1.map.gz emd_9211_half_map_1.map.gz emd_9211_half_map_2.map.gz emd_9211_half_map_2.map.gz | 31.9 MB 59.2 MB 59.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9211 http://ftp.pdbj.org/pub/emdb/structures/EMD-9211 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9211 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9211 | HTTPS FTP |
-Validation report
| Summary document |  emd_9211_validation.pdf.gz emd_9211_validation.pdf.gz | 78.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9211_full_validation.pdf.gz emd_9211_full_validation.pdf.gz | 77.5 KB | Display | |
| Data in XML |  emd_9211_validation.xml.gz emd_9211_validation.xml.gz | 493 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9211 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9211 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9211 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9211 | HTTPS FTP |
-Related structure data
| Related structure data |  9194C  9201C  9202C  9206C  9207C  9208C  9209C  9210C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9211.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9211.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryosparc v0 with curated picks sharpened | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
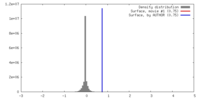
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_9211_msk_1.map emd_9211_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
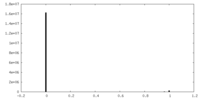
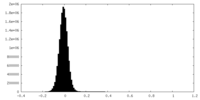
| Density Histograms |
-Additional map: Cryosparc v0 with curated picks unsharpened
| File | emd_9211_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryosparc v0 with curated picks unsharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
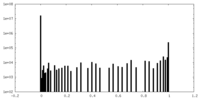
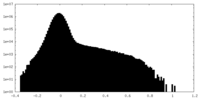
| Density Histograms |
-Half map: Cryosparc v0 with curated picks half map
| File | emd_9211_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryosparc v0 with curated picks half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
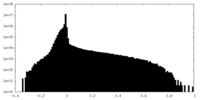
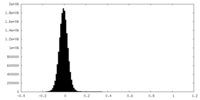
| Density Histograms |
-Half map: Cryosparc v0 with curated picks half map
| File | emd_9211_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryosparc v0 with curated picks half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
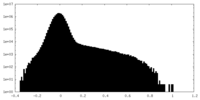
| Density Histograms |
- Sample components
Sample components
-Entire : Rabbit muscle aldolase
| Entire | Name: Rabbit muscle aldolase |
|---|---|
| Components |
|
-Supramolecule #1: Rabbit muscle aldolase
| Supramolecule | Name: Rabbit muscle aldolase / type: complex / ID: 1 / Parent: 0 / Details: Curated picks |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 7420 pixel / Digitization - Dimensions - Height: 7676 pixel / Number grids imaged: 1 / Average exposure time: 6.0 sec. / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)