+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8740 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM reconstruction of PAK pilus from Pseudomonas aeruginosa | |||||||||||||||
 Map data Map data | PAK pilus from Pseudomonas aeruginosa | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||||||||
| Biological species |  Pseudomonas aeruginosa PAK (bacteria) Pseudomonas aeruginosa PAK (bacteria) | |||||||||||||||
| Method | helical reconstruction / cryo EM / Resolution: 8.0 Å | |||||||||||||||
 Authors Authors | Wang F / Osinksi T / Orlova A / Altindal T / Craig L / Egelman EH | |||||||||||||||
| Funding support |  United States, United States,  France, France,  Canada, 4 items Canada, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Structure / Year: 2017 Journal: Structure / Year: 2017Title: Cryoelectron Microscopy Reconstructions of the Pseudomonas aeruginosa and Neisseria gonorrhoeae Type IV Pili at Sub-nanometer Resolution. Authors: Fengbin Wang / Mathieu Coureuil / Tomasz Osinski / Albina Orlova / Tuba Altindal / Gaël Gesbert / Xavier Nassif / Edward H Egelman / Lisa Craig /    Abstract: We report here cryoelectron microscopy reconstructions of type IV pili (T4P) from two important human pathogens, Pseudomonas aeruginosa and Neisseria gonorrhoeae, at ∼ 8 and 5 Å resolution, ...We report here cryoelectron microscopy reconstructions of type IV pili (T4P) from two important human pathogens, Pseudomonas aeruginosa and Neisseria gonorrhoeae, at ∼ 8 and 5 Å resolution, respectively. The two structures reveal distinct arrangements of the pilin globular domains on the pilus surfaces, which impart different helical parameters, but similar packing of the conserved N-terminal α helices, α1, in the filament core. In contrast to the continuous α helix seen in the X-ray crystal structures of the P. aeruginosa and N. gonorrhoeae pilin subunits, α1 in the pilus filaments has a melted segment located between conserved helix-breaking residues Gly14 and Pro22, as seen for the Neisseria meningitidis T4P. Using mutagenesis we show that Pro22 is critical for pilus assembly, as are Thr2 and Glu5, which are positioned to interact in the hydrophobic filament core. These structures provide a framework for understanding T4P assembly, function, and biophysical properties. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8740.map.gz emd_8740.map.gz | 5.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8740-v30.xml emd-8740-v30.xml emd-8740.xml emd-8740.xml | 12.5 KB 12.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8740.png emd_8740.png | 133.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8740 http://ftp.pdbj.org/pub/emdb/structures/EMD-8740 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8740 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8740 | HTTPS FTP |
-Validation report
| Summary document |  emd_8740_validation.pdf.gz emd_8740_validation.pdf.gz | 333.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_8740_full_validation.pdf.gz emd_8740_full_validation.pdf.gz | 332.6 KB | Display | |
| Data in XML |  emd_8740_validation.xml.gz emd_8740_validation.xml.gz | 4.6 KB | Display | |
| Data in CIF |  emd_8740_validation.cif.gz emd_8740_validation.cif.gz | 5.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8740 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8740 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8740 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8740 | HTTPS FTP |
-Related structure data
| Related structure data |  5vxyMC  8739C  5vxxC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8740.map.gz / Format: CCP4 / Size: 13.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8740.map.gz / Format: CCP4 / Size: 13.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PAK pilus from Pseudomonas aeruginosa | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
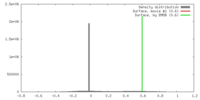
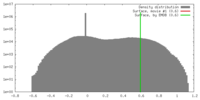
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Pseudomonas aeruginosa Type IV pilin filament
| Entire | Name: Pseudomonas aeruginosa Type IV pilin filament |
|---|---|
| Components |
|
-Supramolecule #1: Pseudomonas aeruginosa Type IV pilin filament
| Supramolecule | Name: Pseudomonas aeruginosa Type IV pilin filament / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAK (bacteria) / Strain: PAK/2pfs Pseudomonas aeruginosa PAK (bacteria) / Strain: PAK/2pfs |
-Macromolecule #1: Fimbrial protein
| Macromolecule | Name: Fimbrial protein / type: protein_or_peptide / ID: 1 / Number of copies: 21 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAK (bacteria) / Strain: PAK/2pfs Pseudomonas aeruginosa PAK (bacteria) / Strain: PAK/2pfs |
| Molecular weight | Theoretical: 15.02118 KDa |
| Sequence | String: FTLIELMIVV AIIGILAAIA IPQYQNYVAR SEGASALASV NPLKTTVEEA LSRGWSVKSG TGTEDATKKE VPLGVAADAN KLGTIALKP DPADGTADIT LTFTMGGAGP KNKGKIITLT RTAADGLWKC TSDQDEQFIP KGCSR |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 9 / Details: 50 mM CHES buffer |
| Grid | Pretreatment - Type: PLASMA CLEANING / Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Average exposure time: 2.0 sec. / Average electron dose: 20.0 e/Å2 Details: Images were stored containing seven parts, where each part represented a set of frames corresponding to a dose of ~20 electrons per Angstrom^2. The full dose image stack was used for the ...Details: Images were stored containing seven parts, where each part represented a set of frames corresponding to a dose of ~20 electrons per Angstrom^2. The full dose image stack was used for the estimation of the CTF as well as for boxing filaments. Only the first two parts were used for the reconstruction (~5 electrons per Angstrom^2) |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 10.5 Å Applied symmetry - Helical parameters - Δ&Phi: 87.3 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Resolution.type: BY AUTHOR / Resolution: 8.0 Å / Resolution method: OTHER / Software - Name: SPIDER / Details: model-map FSC 0.38 cut-off / Number images used: 31231 |
|---|---|
| CTF correction | Software - Name: CTFFIND3 |
| Startup model | Type of model: OTHER / Details: featureless cylinder |
| Final angle assignment | Type: NOT APPLICABLE / Software - Name: SPIDER |
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-5vxy: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)