+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7839 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
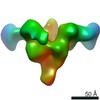
| Title | BBSome complex | |||||||||
 Map data Map data | primary map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.9 Å | |||||||||
 Authors Authors | Chou H / Apelt L / Farrell DP / White SR / Woodsmith J / Svetlov V / Goldstein JS / Nager AR / Li Z / Muller J ...Chou H / Apelt L / Farrell DP / White SR / Woodsmith J / Svetlov V / Goldstein JS / Nager AR / Li Z / Muller J / Dollfus H / Nudler E / Stelzl U / DiMaio F / Walz T / Nachury NV | |||||||||
 Citation Citation |  Journal: Structure / Year: 2019 Journal: Structure / Year: 2019Title: The Molecular Architecture of Native BBSome Obtained by an Integrated Structural Approach. Authors: Hui-Ting Chou / Luise Apelt / Daniel P Farrell / Susan Roehl White / Jonathan Woodsmith / Vladimir Svetlov / Jaclyn S Goldstein / Andrew R Nager / Zixuan Li / Jean Muller / Hélène Dollfus ...Authors: Hui-Ting Chou / Luise Apelt / Daniel P Farrell / Susan Roehl White / Jonathan Woodsmith / Vladimir Svetlov / Jaclyn S Goldstein / Andrew R Nager / Zixuan Li / Jean Muller / Hélène Dollfus / Evgeny Nudler / Ulrich Stelzl / Frank DiMaio / Maxence V Nachury / Thomas Walz /    Abstract: The unique membrane composition of cilia is maintained by a diffusion barrier at the transition zone that is breached when the BBSome escorts signaling receptors out of cilia. Understanding how the ...The unique membrane composition of cilia is maintained by a diffusion barrier at the transition zone that is breached when the BBSome escorts signaling receptors out of cilia. Understanding how the BBSome removes proteins from cilia has been hampered by a lack of structural information. Here, we present a nearly complete Cα model of BBSome purified from cow retina. The model is based on a single-particle cryo-electron microscopy density map at 4.9-Å resolution that was interpreted with the help of comprehensive Rosetta-based structural modeling constrained by crosslinking mass spectrometry data. We find that BBSome subunits have a very high degree of interconnectivity, explaining the obligate nature of the complex. Furthermore, like other coat adaptors, the BBSome exists in an autoinhibited state in solution and must thus undergo a conformational change upon recruitment to membranes by the small GTPase ARL6/BBS3. Our model provides the first detailed view of the machinery enabling ciliary exit. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7839.map.gz emd_7839.map.gz | 11.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7839-v30.xml emd-7839-v30.xml emd-7839.xml emd-7839.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7839_fsc.xml emd_7839_fsc.xml | 11 KB | Display |  FSC data file FSC data file |
| Images |  emd_7839.png emd_7839.png | 54.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7839 http://ftp.pdbj.org/pub/emdb/structures/EMD-7839 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7839 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7839 | HTTPS FTP |
-Validation report
| Summary document |  emd_7839_validation.pdf.gz emd_7839_validation.pdf.gz | 78.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_7839_full_validation.pdf.gz emd_7839_full_validation.pdf.gz | 78 KB | Display | |
| Data in XML |  emd_7839_validation.xml.gz emd_7839_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7839 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7839 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7839 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7839 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7839.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7839.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | primary map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.23 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : BBSome
| Entire | Name: BBSome |
|---|---|
| Components |
|
-Supramolecule #1: BBSome
| Supramolecule | Name: BBSome / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: BBS5
| Macromolecule | Name: BBS5 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MSVLDALWED RDVRFDVSSQ QMKTRPGEVL IDCLDSVEDT KGNNGDRGRL LVTNLRIVWH SLALPRVNL SIGYNCILNI TTRTANSKLR GQTEALYVLT KCNSTRFEFI FTNLVPGSPR L YTSLIAVH RAYETSKMYR DFKLRSALIQ NKQLRLLPQE NVYNKINGVW ...String: MSVLDALWED RDVRFDVSSQ QMKTRPGEVL IDCLDSVEDT KGNNGDRGRL LVTNLRIVWH SLALPRVNL SIGYNCILNI TTRTANSKLR GQTEALYVLT KCNSTRFEFI FTNLVPGSPR L YTSLIAVH RAYETSKMYR DFKLRSALIQ NKQLRLLPQE NVYNKINGVW NLSSDQGNLG TF FITNVRI VWHANMNDSF NVSIPYLQIR SVKIRDSKFG LALVIESSQQ SGGYVLGFKI DPV EKLQES VKEINSLHKV YSANPIFGVD YEMEEKPQPL EALTVKQIQD DVEIDSDDHT DAFV AYFAD GNKQQDREPV FSEELGLAIE KLKDGFTLQG LWEVMN |
-Macromolecule #2: BBS9
| Macromolecule | Name: BBS9 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MSLFKARDWW STVLGDKEEF DQGCLCLADV DNTGNGQDKI IVGSFMGYLR IFNPHPVKTG DGAQAEDLL LEVHLRDPIL QVEVGKFVSG TEMLHLAVLH SRKLCVYSVS GTLGNVEHGN Q YQIKLMYE HNLQRTACNM TYGSFGGVKG RDLICIQSVD GMLMVFEQES ...String: MSLFKARDWW STVLGDKEEF DQGCLCLADV DNTGNGQDKI IVGSFMGYLR IFNPHPVKTG DGAQAEDLL LEVHLRDPIL QVEVGKFVSG TEMLHLAVLH SRKLCVYSVS GTLGNVEHGN Q YQIKLMYE HNLQRTACNM TYGSFGGVKG RDLICIQSVD GMLMVFEQES YAFGRFLPGS LL PGPLAYS SRTDSFITVS SCHQVESYKY QVLAFATDAD KRQETEQQKH GSGKRLVVDW TLN IGEQAI DICIVSFIQS ASSVFVLGER NFFCLKDNGQ IQFMKKLDYS PSCFLPYCSV SEGT INTLI GNHNNMLHIY QDVTLKWATQ LPHVPVAVRV GCLHDLKGVI VTLSDDGHLQ CSYLG TDPS LFQAPKVESR ELNYDELDME LKELQKVIKN VNKSQDVWPL TEREDDLKVS AMVSPN FDS VSQATDVEVG ADLVPSVTVK VTLKNRVALQ KIKLSIYVQP PLVLTGDQFT FEFMAPE MT RTVGFSVYLK GSYSPPELEG NAVVSYSRPT ERNPDGIPRV SQCKFRLPLK LVCLPGQP S KTASHKLTID TNKSPVSLLS LFPGFAKQSE DDQVNVMGFR FLGGSQVTLL ASKTSQRYR IQSEQFEDLW LITNELIIRL QEYFEKQGIK DFTCSFSGSV PLEEYFELID HHFELRINGE KLEELLSER AVQFRAIQRR LLTRFKDKTP APLQHLDTLL DGTYKQVIAL ADAVEENQDN L FQSFTRLK SATHLVILLI GLWQKLSADQ IAILEAAFLP LQQDTQELGW EETVDAALSH LL KTCLSKS SKEQALNLNS QLGIPKDTSQ LKKHITLFCD RLAKGGRLCL STDAAAPQTM VMP GGCATI PESDLEGRSI DQDSSELFTN HKHLMVETPV PEVSPLQGVT E |
-Macromolecule #3: BBS1
| Macromolecule | Name: BBS1 / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MAATSSSDSD GGKGESSEAN SKWLDSLSDS MANIHTFSAC LALADFHGDG EYKLAMGDLG PDGRQPRLK VLKGHTLVSQ KPLPDLPAAA VTFLMASHEP RTPALAIASG PCVYVYKNLK P YFKFSLPS LPTNPLEQDL WNQAKEDQID PLTLKEMLEG IREKAEVPLS ...String: MAATSSSDSD GGKGESSEAN SKWLDSLSDS MANIHTFSAC LALADFHGDG EYKLAMGDLG PDGRQPRLK VLKGHTLVSQ KPLPDLPAAA VTFLMASHEP RTPALAIASG PCVYVYKNLK P YFKFSLPS LPTNPLEQDL WNQAKEDQID PLTLKEMLEG IREKAEVPLS VQSLRFLPLE LS EMEAFVN QHKSKSIRRQ TVITTMTTLK KNLADEDAVS CLVLGTENKE LLVLDPEAFT ILA KMSLPS VPAFLEASGQ FDVEFRLAAA CRNGSIYILR RDSKRPKYCI ELGAQPVGLV GVHK VLVVG SNQDSLHGFT YKGKRLWTVQ MPAAILAMNL LEQHSRGLQA VMAALANEEV RIYHD KVLL NVIRTPEAVT SLCFGRYGRE DNTLIMTTLG GGLIIKILKR TAVFAEGGGE AGPPPS QAI KLNVPRKTRL YVDQTLRERE AGTAMHRTFQ ADLYLLRLRA ARAYVQALES SLSPVSL TA REPLKLHAVV QGLGPTFKLT LHLQNTSTAR PILGLVVCFL YNEVLYALPR AFFKVPLL V PGLNYPLETF VKSLSDKGIS DIIKVLVLRE GQSTPLLSAH INMPMSEGLA AD |
-Macromolecule #4: BBS7
| Macromolecule | Name: BBS7 / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MLFSSSVPFV GVTSQKTMKL LPASKHRATQ KVVVGDHDGI VMCFGMKKGE AVTVFKTLPG QKIARLELG GALNTPQEKI FIAAGSEIRG FTKRGKQFLS FETNLTESIK AMHISGSDLF L SASYIYNH YCDCKDQHYY LSGDKINDVI CLPVERLLRE VPVLACQDRV ...String: MLFSSSVPFV GVTSQKTMKL LPASKHRATQ KVVVGDHDGI VMCFGMKKGE AVTVFKTLPG QKIARLELG GALNTPQEKI FIAAGSEIRG FTKRGKQFLS FETNLTESIK AMHISGSDLF L SASYIYNH YCDCKDQHYY LSGDKINDVI CLPVERLLRE VPVLACQDRV LRVLQGSDVT YE IEVPGPP TVLALHNGNG GDSGEDLLFG TSDGKLGLIQ ITTSKPIHKW EIRNEKKRGG ILC VDSFDI VGDGVKDLLV GRDDGMVEVY GFDNANEPVL RFDHTLSESV TSIQGGCVGK DGYD EIVVS TYSGWITGLT TEPVHKESGP GEELKFNQEM QNKISSLRSE LEQLQYKVLQ EREKY QQSS QSSKAKSAVP SFSVNDKFTL NKDDASYSLI LEVQTAIDNV LIQSDVPIDL LDVDKN SAV VSFSSCDSES NDNFLLATYR CQANTTRLEL KIRSIEGQYG TLQAYVTPRI QPKTCQV RQ YHIKPLSLHQ RTHFIDHDRP MNTLTLTGQF SFSELHSWVV FCMPEVPEKP PAGECVTF Y FQNTFLDTQL ESTYRKGEGV FKSDNISTIS ILKDVLSKEA TKRKINLNIS YEINEVSVK HTLKLIHPKL EYQLLLAKKV QLIDALKELQ VHEGNTNFLI PEYRCILEEA DHLQEEYKKQ PAHLERLYG MITDLFIDKF KFKGTNVKTK VPLLLEILDS YDQNALIAFF DAA |
-Macromolecule #5: BBS8
| Macromolecule | Name: BBS8 / type: protein_or_peptide / ID: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: APASDMEPLL LAWSYFRRRR FQLCADLCTQ MLEKSPCDQA AWILKARALT EMVYVDEIDV DEEGIAEMI LDENAIAQVP RPGTSLKLPG TNQTGGPSPA VRPVTQAGRP ITGFLRPSTQ S GRPGTIEQ AIKTPRTAYT ARPIASSSGR FVRLGTASML TSPDGPFINL ...String: APASDMEPLL LAWSYFRRRR FQLCADLCTQ MLEKSPCDQA AWILKARALT EMVYVDEIDV DEEGIAEMI LDENAIAQVP RPGTSLKLPG TNQTGGPSPA VRPVTQAGRP ITGFLRPSTQ S GRPGTIEQ AIKTPRTAYT ARPIASSSGR FVRLGTASML TSPDGPFINL SRLNLAKYAQ KP KLAKALF EYIFHHENDV KTALDLAALS TEHSQYKDWW WKVQIGKCYY RLGLYREAEK QFK SALKQQ EMVDTFLYLA KVYISLDQPL TALNLFKQGL DKFPGEVTLL CGIARIYEEM NNIS SATEY YKEVLKQDNT HVEAIACIGS NHFYTDQPEV ALRFYRRLLQ MGVYNCQLFN NLGLC CFYA QQYDMTLTSF ERALSLAENE EEVADVWYNL GHVAVGTGDT NLAHQCFRLA LVSNNQ HAE AYNNLAVLEM RRGHVEQAKA LLQTASSLAP HMYEPHFNFA TISDKIGDLQ RSYAAAK KS EAAFPDHVDT QHLIKQLEQH FAML |
-Macromolecule #6: BBS18
| Macromolecule | Name: BBS18 / type: protein_or_peptide / ID: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MLSAAHKSFC FPSATQDETV SNISDMAETK SMFREVLPKQ DLGQLYVEDI TTMVLCKPKL LPLKSLTLE KLEKMQQAAQ DTIHQQEMTE KEQKITH |
-Macromolecule #7: BBS4
| Macromolecule | Name: BBS4 / type: protein_or_peptide / ID: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MAEEKLSART QLPVSAESQK PVLKKAPEFP ILEKQNWLIH LYYIQKDYEA CKAVIKEQLQ ETHGLCEYA IYVQALIFRL EGNIQESLRL FQMCAFLSPQ CADNLKQVAR SLFLLGKHKA A IEVYNEAA KLNQKDWEIC HNLGVCYIYL KQFDKAQDQL HNALHLNRHD ...String: MAEEKLSART QLPVSAESQK PVLKKAPEFP ILEKQNWLIH LYYIQKDYEA CKAVIKEQLQ ETHGLCEYA IYVQALIFRL EGNIQESLRL FQMCAFLSPQ CADNLKQVAR SLFLLGKHKA A IEVYNEAA KLNQKDWEIC HNLGVCYIYL KQFDKAQDQL HNALHLNRHD LTYIMLGKIF LL KGDLDKA IEIYKKAVEF SPENTELLTT LGLLYLQLGI YQKAFEHLGN TLTYDPTNYK AIL AAGSMM QTHGDFDVAL TKYKVVACAV IESPPLWNNI GMCFFGKKKY VAAISCLKRA NYLA PLDWK ILYNLGLVHL TMQQYASAFH FLSAAINFQP KMGELYMLLA VALTNLEDSE NAKRA YEEA VRLDKCNPLV NLNYAVLLYN QGEKRDALAQ YQEMEKKVNL LKYSSSLEFD PEMVEV AQK LGAALQVGEA LVWTKPVKDP KSKHQTASTS KAAGFQQPLG SNQALGQAMS SAATCRK LS SGAGGTSQLT KPPSLPLEPE PTVEAQPTEA SAQTREK |
-Macromolecule #8: BBS2
| Macromolecule | Name: BBS2 / type: protein_or_peptide / ID: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MLQPVFTLKL RHKISPRMVA VGRYDGTHPC LAAATQAGKV FIHNPHSRSQ HLGAPRVLQS PLESDVSLL NINQTVSCLT AGVLNPELGY DALLVGTQTN LLAYDVYNNS DLFYREVADG A SAIVLGTL GDITSPLAII GGNCALQGFN HEGNDLFWTV TGDNVHSLAL ...String: MLQPVFTLKL RHKISPRMVA VGRYDGTHPC LAAATQAGKV FIHNPHSRSQ HLGAPRVLQS PLESDVSLL NINQTVSCLT AGVLNPELGY DALLVGTQTN LLAYDVYNNS DLFYREVADG A SAIVLGTL GDITSPLAII GGNCALQGFN HEGNDLFWTV TGDNVHSLAL CDFDGDGKKE LL VGSEDFD IRVFKEDEIV AEMSETEIIT SLCPMYGSRF GYALSNGTVG VYDKTARYWR IKS KNQAMS IHAFDLNSDG VCELITGWSN GKVDARSDRT GEVIFKDNFS SAIAGVVEGD YRME GCQQL ICCSVDGEIR GYLPGTAEMR GNLMDISVEQ DLIRELSQKK QNLLLELRNY EENAK AELS SPLNEADGHR GVIPANTKHH TALSVSLGSE AQAAHAELCI STSNDTIIRA VLIFAE GVF AGESHVVHPS VHHLSSSVRI PITPPKDIPV DLHLKTFVGY RSSTQFHVFE LTRQLPR FS MYALTSPDPA SEPLSYVNFI IAERAQRVVM WLNQNFLLPE DTNIQNAPFQ VCFTSLRN G GQLYIKIKLS GEITVNTDDI DLAGDIIQSM ASFFAIEDLQ VEADFPVYFE ELRKVLVKV DEYHSVHQKL SADMADNSNL IRSLLVQAED ARLMRDMKTM KNRYKELYDL NKDLLNGYKI RCNNHTELL GSLKAVNQAI QRAGHLRVGK PKNQVITACR DAIRSNNINM LFRIMRVGTA S S |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 / Details: 40 mM HEPES, 220 mM NaCl, 1 mM DTT |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: GATAN K2 SUMMIT (4k x 4k) / #0 - Detector mode: SUPER-RESOLUTION / #0 - Average electron dose: 46.8 e/Å2 / #1 - Image recording ID: 2 / #1 - Film or detector model: GATAN K2 SUMMIT (4k x 4k) / #1 - Detector mode: SUPER-RESOLUTION / #1 - Average electron dose: 89.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















