[English] 日本語
 Yorodumi
Yorodumi- EMDB-7818: Human nuclear exosome-MTR4 RNA complex - overall reconstruction -... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7818 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human nuclear exosome-MTR4 RNA complex - overall reconstruction - small class KOW domain open | |||||||||
 Map data Map data | Overall reconstruction for small class of human nuclear exosome-MTR4 complex with the MTR4 KOW domain in open conformation | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.75 Å | |||||||||
 Authors Authors | Weick E-M / Lima CD | |||||||||
 Citation Citation |  Journal: Cell / Year: 2018 Journal: Cell / Year: 2018Title: Helicase-Dependent RNA Decay Illuminated by a Cryo-EM Structure of a Human Nuclear RNA Exosome-MTR4 Complex. Authors: Eva-Maria Weick / M Rhyan Puno / Kurt Januszyk / John C Zinder / Michael A DiMattia / Christopher D Lima /  Abstract: The ribonucleolytic RNA exosome interacts with RNA helicases to degrade RNA. To understand how the 3' to 5' Mtr4 helicase engages RNA and the nuclear exosome, we reconstituted 14-subunit Mtr4- ...The ribonucleolytic RNA exosome interacts with RNA helicases to degrade RNA. To understand how the 3' to 5' Mtr4 helicase engages RNA and the nuclear exosome, we reconstituted 14-subunit Mtr4-containing RNA exosomes from Saccharomyces cerevisiae, Schizosaccharomyces pombe, and human and show that they unwind structured substrates to promote degradation. We loaded a human exosome with an optimized DNA-RNA chimera that stalls MTR4 during unwinding and determined its structure to an overall resolution of 3.45 Å by cryoelectron microscopy (cryo-EM). The structure reveals an RNA-engaged helicase atop the non-catalytic core, with RNA captured within the central channel and DIS3 exoribonuclease active site. MPP6 tethers MTR4 to the exosome through contacts to the RecA domains of MTR4. EXOSC10 remains bound to the core, but its catalytic module and cofactor C1D are displaced by RNA-engaged MTR4. Competition for the exosome core may ensure that RNA is committed to degradation by DIS3 when engaged by MTR4. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7818.map.gz emd_7818.map.gz | 13.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7818-v30.xml emd-7818-v30.xml emd-7818.xml emd-7818.xml | 32.6 KB 32.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7818_fsc.xml emd_7818_fsc.xml | 13.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_7818.png emd_7818.png | 49.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7818 http://ftp.pdbj.org/pub/emdb/structures/EMD-7818 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7818 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7818 | HTTPS FTP |
-Validation report
| Summary document |  emd_7818_validation.pdf.gz emd_7818_validation.pdf.gz | 78 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_7818_full_validation.pdf.gz emd_7818_full_validation.pdf.gz | 77.1 KB | Display | |
| Data in XML |  emd_7818_validation.xml.gz emd_7818_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7818 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7818 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7818 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7818 | HTTPS FTP |
-Related structure data
| Related structure data |  7808C  7809C  7810C  7812C  7813C  7814C  7815C  7819C  6d6qC  6d6rC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7818.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7818.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Overall reconstruction for small class of human nuclear exosome-MTR4 complex with the MTR4 KOW domain in open conformation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
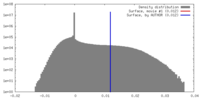
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Human nuclear exosome-MTR4 helicase captured after unwinding a DN...
+Supramolecule #1: Human nuclear exosome-MTR4 helicase captured after unwinding a DN...
+Macromolecule #1: EXOSC9
+Macromolecule #2: EXOSC4
+Macromolecule #3: EXOSC8
+Macromolecule #4: EXOSC5
+Macromolecule #5: EXOSC7
+Macromolecule #6: EXOSC6
+Macromolecule #7: EXOSC3
+Macromolecule #8: EXOSC2
+Macromolecule #9: EXOSC1
+Macromolecule #10: EXOSC10
+Macromolecule #11: DIS3
+Macromolecule #12: MPP6, MPHOSPH6
+Macromolecule #13: MTR4, MTREX
+Macromolecule #14: C1D, Rrp47
+Macromolecule #15: synthetic RNA
+Macromolecule #16: DNA-RNA chimera
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: GOLD / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.037 kPa | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV / Details: 30 sec wait time, 2.5 sec blot time. | |||||||||||||||||||||
| Details | Sample was monodisperse upon elution from gel filtration prior to vitrification. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number real images: 1439 / Average electron dose: 85.23 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | Models can be manually docked into this map. |
|---|
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)