[English] 日本語
 Yorodumi
Yorodumi- EMDB-7312: Structure of human PRC2 bound to a hetero-dinucleosome substrate ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7312 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human PRC2 bound to a hetero-dinucleosome substrate with 35 bp linker DNA. Refinement after signal subtraction of the modified nucleosome ton improve resolution of the substrate nucleosome - PRC2 interface. | |||||||||
 Map data Map data | Structure of human PRC2 bound to a hetero-dinucleosome substrate with 30 bp linker DNA. Refinement after signal subtraction of the modified nucleosome. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
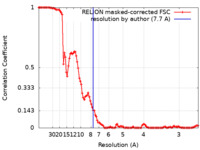
| Method | single particle reconstruction / cryo EM / Resolution: 7.7 Å | |||||||||
 Authors Authors | Poepsel S / Kasinath V / Nogales E | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2018 Journal: Nat Struct Mol Biol / Year: 2018Title: Cryo-EM structures of PRC2 simultaneously engaged with two functionally distinct nucleosomes. Authors: Simon Poepsel / Vignesh Kasinath / Eva Nogales /  Abstract: Epigenetic regulation is mediated by protein complexes that couple recognition of chromatin marks to activity or recruitment of chromatin-modifying enzymes. Polycomb repressive complex 2 (PRC2), a ...Epigenetic regulation is mediated by protein complexes that couple recognition of chromatin marks to activity or recruitment of chromatin-modifying enzymes. Polycomb repressive complex 2 (PRC2), a gene silencer that methylates lysine 27 of histone H3, is stimulated upon recognition of its own catalytic product and has been shown to be more active on dinucleosomes than H3 tails or single nucleosomes. These properties probably facilitate local H3K27me2/3 spreading, causing heterochromatin formation and gene repression. Here, cryo-EM reconstructions of human PRC2 bound to bifunctional dinucleosomes show how a single PRC2, via interactions with nucleosomal DNA, positions the H3 tails of the activating and substrate nucleosome to interact with the EED subunit and the SET domain of EZH2, respectively. We show how the geometry of the PRC2-DNA interactions allows PRC2 to accommodate varying lengths of the linker DNA between nucleosomes. Our structures illustrate how an epigenetic regulator engages with a complex chromatin substrate. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7312.map.gz emd_7312.map.gz | 116.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7312-v30.xml emd-7312-v30.xml emd-7312.xml emd-7312.xml | 10 KB 10 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7312_fsc.xml emd_7312_fsc.xml | 11.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_7312.png emd_7312.png | 40.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7312 http://ftp.pdbj.org/pub/emdb/structures/EMD-7312 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7312 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7312 | HTTPS FTP |
-Validation report
| Summary document |  emd_7312_validation.pdf.gz emd_7312_validation.pdf.gz | 375.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_7312_full_validation.pdf.gz emd_7312_full_validation.pdf.gz | 375.2 KB | Display | |
| Data in XML |  emd_7312_validation.xml.gz emd_7312_validation.xml.gz | 11.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7312 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7312 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7312 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7312 | HTTPS FTP |
-Related structure data
| Related structure data |  7306C  7307C  7308C  7309C  7310C  7311C  7313C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7312.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7312.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of human PRC2 bound to a hetero-dinucleosome substrate with 30 bp linker DNA. Refinement after signal subtraction of the modified nucleosome. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.32 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
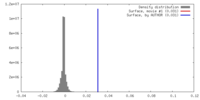
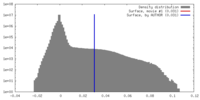
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human Polycomb Repressive Complex 2 (PRC2) in complex with a hete...
| Entire | Name: Human Polycomb Repressive Complex 2 (PRC2) in complex with a heterodinucleosome composed of an unmodified nucleosome and a nucleosome carrying an H3K27 pseudo-trimethyl mark, connected by 30 bp of linker DNA |
|---|---|
| Components |
|
-Supramolecule #1: Human Polycomb Repressive Complex 2 (PRC2) in complex with a hete...
| Supramolecule | Name: Human Polycomb Repressive Complex 2 (PRC2) in complex with a heterodinucleosome composed of an unmodified nucleosome and a nucleosome carrying an H3K27 pseudo-trimethyl mark, connected by 30 bp of linker DNA type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.9 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)