+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7137 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | PRMT5:MEP50 complex | |||||||||
 Map data Map data | PRMT5:MEP50 complex | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
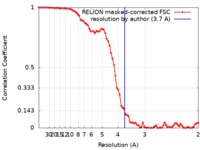
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Timm DE | |||||||||
 Citation Citation |  Journal: PLoS One / Year: 2018 Journal: PLoS One / Year: 2018Title: Cryo-electron microscopy structure of a human PRMT5:MEP50 complex. Authors: David E Timm / Valorie Bowman / Russell Madsen / Charles Rauch /  Abstract: Protein arginine methyl transferase 5 (PRMT5) is a signaling protein and histone modifying enzyme that is important in many cellular processes, including regulation of eukaryotic gene transcription. ...Protein arginine methyl transferase 5 (PRMT5) is a signaling protein and histone modifying enzyme that is important in many cellular processes, including regulation of eukaryotic gene transcription. Reported here is a 3.7 Å structure of PRMT5, solved in complex with regulatory binding subunit MEP50 (methylosome associated protein 50, WDR77, p44), by single particle (SP) cryo-Electron Microscopy (cryo-EM) using micrographs of particles that are visibly crowded and aggregated. Despite suboptimal micrograph appearance, this cryo-EM structure is in good agreement with previously reported crystal structures of the complex, which revealed a 450 kDa hetero-octameric assembly having internal D2 symmetry. The catalytic PRMT5 subunits form a core tetramer and the MEP50 subunits are arranged peripherally in complex with the PRMT5 N-terminal domain. The cryo-EM reconstruction shows good side chain definition and shows a well-resolved peak for a bound dehydrosinefungin inhibitor molecule. These results demonstrate the applicability of cryo-EM in determining structures of human protein complexes of biomedical significance and suggests cryo-EM could be further utilized to understand PRMT5 interactions with other biologically important binding proteins and ligands. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7137.map.gz emd_7137.map.gz | 38 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7137-v30.xml emd-7137-v30.xml emd-7137.xml emd-7137.xml | 12.2 KB 12.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7137_fsc.xml emd_7137_fsc.xml | 7.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_7137.png emd_7137.png | 176.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7137 http://ftp.pdbj.org/pub/emdb/structures/EMD-7137 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7137 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7137 | HTTPS FTP |
-Validation report
| Summary document |  emd_7137_validation.pdf.gz emd_7137_validation.pdf.gz | 77.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_7137_full_validation.pdf.gz emd_7137_full_validation.pdf.gz | 76.5 KB | Display | |
| Data in XML |  emd_7137_validation.xml.gz emd_7137_validation.xml.gz | 493 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7137 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7137 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7137 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7137 | HTTPS FTP |
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7137.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7137.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PRMT5:MEP50 complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : PRMT5:MEP50
| Entire | Name: PRMT5:MEP50 |
|---|---|
| Components |
|
-Supramolecule #1: PRMT5:MEP50
| Supramolecule | Name: PRMT5:MEP50 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: The complex contains a tetramer of PRMT5:MEP50 heterodimers |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Molecular weight | Experimental: 450 KDa |
-Macromolecule #1: Protein arginine N-methyltransferase 5
| Macromolecule | Name: Protein arginine N-methyltransferase 5 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MRGPNSGTEK GRLVIPEKQG FDFLCMPVFH PRFKREFIQE PAKNRPGPQT RSDLLLSGRA FLLPLNQEDN TNLARVLTN HIHTGHHSSM FWMRVPLVAP EDLRDDIIEN APTTHTEEYS GEEKTWMWWH NFRTLCDYSK R IAVALEIG ADLPSNHVID RWLGEPIKAA ...String: MRGPNSGTEK GRLVIPEKQG FDFLCMPVFH PRFKREFIQE PAKNRPGPQT RSDLLLSGRA FLLPLNQEDN TNLARVLTN HIHTGHHSSM FWMRVPLVAP EDLRDDIIEN APTTHTEEYS GEEKTWMWWH NFRTLCDYSK R IAVALEIG ADLPSNHVID RWLGEPIKAA ILPTSIFLTN KKGFPVLSKM HQRLIFRLLK LEVQFIITGT NH HSEKEFC SYLQYLEYLS QNRPPPNAYE LFAKGYEDYL QSPLQPLMDN LESQTYEVFE KDPIKYSQYQ QAI YKCLLD RVPEEEKDTN VQVLMVLGAG RGPLVNASLR AAKQADRRIK LYAVEKNPNA VVTLENWQFE EWGS QVTVV SSDMREWVAP EKADIIVSEL LGSFADNELS PECLDGAQHF LKDDGVSIPG EYTSFLAPIS SSKLY NEVR ACREKDRDPE AQFEMPYVVR LHNFHQLSAP QPCFTFSHPN RDPMIDNNRY CTLEFPVEVN TVLHGF AGY FETVLYQDIT LSIRPETHSP GMFSWFPILF PIKQPITVRE GQTICVRFWR CSNSKKVWYE WAVTAPV CS AIHNPTGRSY TIGL |
-Macromolecule #2: Methylosome protein 50
| Macromolecule | Name: Methylosome protein 50 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MRKETPPPLV PPAAREWNLP PNAPACMERQ LEAARYRSDG ALLLGASSLS GRCWAGSLWL FKDPCAAPNE GFCSAGVQT EAGVADLTWV GERGILVASD SAHAAQVTCV AASPHKDSVF LSCSEDNRIL LWDTRCPKPA S QIGCSAPG YLPTSLAWHP QQSEVFVFGD ...String: MRKETPPPLV PPAAREWNLP PNAPACMERQ LEAARYRSDG ALLLGASSLS GRCWAGSLWL FKDPCAAPNE GFCSAGVQT EAGVADLTWV GERGILVASD SAHAAQVTCV AASPHKDSVF LSCSEDNRIL LWDTRCPKPA S QIGCSAPG YLPTSLAWHP QQSEVFVFGD ENGTVSLVDT KSTSCVLSSA VHSQCVTGLV FSPHSVPFLA SL SEDCSLA VLDSSLSELF RSQAHRDFVR DATWSPLNHS LLTTVGWDHQ VVHHVVPTEP LPAPGPASVT E |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.38 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 50 mM HEPES, 150 mM NaCl |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Instrument: GATAN CRYOPLUNGE 3 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 193 / Average exposure time: 8.0 sec. / Average electron dose: 64.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)