[English] 日本語
 Yorodumi
Yorodumi- EMDB-6998: Cryo-EM structure of the human alpha5beta3 GABAA receptor in comp... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6998 | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human alpha5beta3 GABAA receptor in complex with GABA and Nb25 | ||||||||||||||||||||||||||||||||||||||||||
 Map data Map data | CryoEM Structure of the human %u03B15%u03B23 GABAA receptor | ||||||||||||||||||||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||||||||||||||||||||
 Keywords Keywords | IRON CHANNEL / MEMBRANE PROTEIN | ||||||||||||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationinhibitory extracellular ligand-gated monoatomic ion channel activity / benzodiazepine receptor activity / cellular response to histamine / GABA receptor activation / inner ear receptor cell development / GABA-gated chloride ion channel activity / GABA-A receptor activity / GABA-A receptor complex / GABA receptor binding / inhibitory synapse assembly ...inhibitory extracellular ligand-gated monoatomic ion channel activity / benzodiazepine receptor activity / cellular response to histamine / GABA receptor activation / inner ear receptor cell development / GABA-gated chloride ion channel activity / GABA-A receptor activity / GABA-A receptor complex / GABA receptor binding / inhibitory synapse assembly / innervation / synaptic transmission, GABAergic / postsynaptic specialization membrane / gamma-aminobutyric acid signaling pathway / nervous system process / neurotransmitter receptor activity / cochlea development / neuronal cell body membrane / roof of mouth development / Signaling by ERBB4 / associative learning / chloride channel complex / GABA-ergic synapse / transmembrane transporter complex / behavioral fear response / regulation of postsynaptic membrane potential / chloride transmembrane transport / dendrite membrane / ligand-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / regulation of membrane potential / cytoplasmic vesicle membrane / presynaptic membrane / signaling receptor activity / postsynapse / postsynaptic membrane / neuron projection / synapse / signal transduction / nucleoplasm / identical protein binding / plasma membrane / cytosol Similarity search - Function | ||||||||||||||||||||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | ||||||||||||||||||||||||||||||||||||||||||
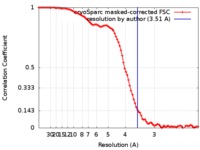
| Method | single particle reconstruction / cryo EM / Resolution: 3.51 Å | ||||||||||||||||||||||||||||||||||||||||||
 Authors Authors | Liu S / Xu L | ||||||||||||||||||||||||||||||||||||||||||
| Funding support |  China, China,  United States, 13 items United States, 13 items
| ||||||||||||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Cell Res / Year: 2018 Journal: Cell Res / Year: 2018Title: Cryo-EM structure of the human α5β3 GABA receptor. Authors: Si Liu / Lingyi Xu / Fenghui Guan / Yun-Tao Liu / Yanxiang Cui / Qing Zhang / Xiang Zheng / Guo-Qiang Bi / Z Hong Zhou / Xiaokang Zhang / Sheng Ye /   | ||||||||||||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6998.map.gz emd_6998.map.gz | 27.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6998-v30.xml emd-6998-v30.xml emd-6998.xml emd-6998.xml | 16.4 KB 16.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_6998_fsc.xml emd_6998_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_6998.png emd_6998.png | 144.4 KB | ||
| Filedesc metadata |  emd-6998.cif.gz emd-6998.cif.gz | 6.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6998 http://ftp.pdbj.org/pub/emdb/structures/EMD-6998 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6998 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6998 | HTTPS FTP |
-Validation report
| Summary document |  emd_6998_validation.pdf.gz emd_6998_validation.pdf.gz | 453.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_6998_full_validation.pdf.gz emd_6998_full_validation.pdf.gz | 453.2 KB | Display | |
| Data in XML |  emd_6998_validation.xml.gz emd_6998_validation.xml.gz | 11.3 KB | Display | |
| Data in CIF |  emd_6998_validation.cif.gz emd_6998_validation.cif.gz | 14.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6998 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6998 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6998 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6998 | HTTPS FTP |
-Related structure data
| Related structure data |  6a96MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6998.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6998.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM Structure of the human %u03B15%u03B23 GABAA receptor | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
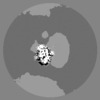
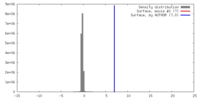
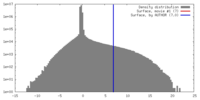
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Pentameric complex of 1 alpha and 4 beta GABA receptor
| Entire | Name: Pentameric complex of 1 alpha and 4 beta GABA receptor |
|---|---|
| Components |
|
-Supramolecule #1: Pentameric complex of 1 alpha and 4 beta GABA receptor
| Supramolecule | Name: Pentameric complex of 1 alpha and 4 beta GABA receptor type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Nb25
| Macromolecule | Name: Nb25 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.673024 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLQESGGG LVQAGGSLRL SCAASGHTFN YPIMGWFRQA PGKEREFVGA ISWSGGSTSY ADSVKDRFTI SRDNAKNTVY LEMNNLKPE DTAVYYCAAK GRYSGGLYYP TNYDYWGQGT QVTVSS |
-Macromolecule #2: Gamma-aminobutyric acid receptor subunit beta-3,Gamma-aminobutyri...
| Macromolecule | Name: Gamma-aminobutyric acid receptor subunit beta-3,Gamma-aminobutyric acid receptor subunit beta-3 type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 42.052711 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MWGLAGGRLF GIFSAPVLVA VVCCAQSVND PGNMSFVKET VDKLLKGYDI RLRPDFGGPP VCVGMNIDIA SIDMVSEVNM DYTLTMYFQ QYWRDKRLAY SGIPLNLTLD NRVADQLWVP DTYFLNDKKS FVHGVTVKNR MIRLHPDGTV LYGLRITTTA A CMMDLRRY ...String: MWGLAGGRLF GIFSAPVLVA VVCCAQSVND PGNMSFVKET VDKLLKGYDI RLRPDFGGPP VCVGMNIDIA SIDMVSEVNM DYTLTMYFQ QYWRDKRLAY SGIPLNLTLD NRVADQLWVP DTYFLNDKKS FVHGVTVKNR MIRLHPDGTV LYGLRITTTA A CMMDLRRY PLDEQNCTLE IESYGYTTDD IEFYWRGGDK AVTGVERIEL PQFSIVEHRL VSRNVVFATG AYPRLSLSFR LK RNIGYFI LQTYMPSILI TILSWVSFWI NYDASAARVA LGITTVLTMT TINTHLRETL PKIPYVKAID MYLMGCFVFV FLA LLEYAF VNYIFFSQPA RAAAIDRWSR IVFPFTFSLF NLVYWLYYVN UniProtKB: Gamma-aminobutyric acid receptor subunit beta-3, Gamma-aminobutyric acid receptor subunit beta-3 |
-Macromolecule #3: Gamma-aminobutyric acid receptor subunit alpha-5,Gamma-aminobutyr...
| Macromolecule | Name: Gamma-aminobutyric acid receptor subunit alpha-5,Gamma-aminobutyric acid receptor subunit alpha-5 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 44.456113 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDNGMFSGFI MIKNLLLFCI SMNLSSHFGF SQMPTSSVKD ETNDNITIFT RILDGLLDGY DNRLRPGLGE RITQVRTDIY VTSFGPVSD TEMEYTIDVF FRQSWKDERL RFKGPMQRLP LNNLLASKIW TPDTFFHNGK KSIAHNMTTP NKLLRLEDDG T LLYTMRLT ...String: MDNGMFSGFI MIKNLLLFCI SMNLSSHFGF SQMPTSSVKD ETNDNITIFT RILDGLLDGY DNRLRPGLGE RITQVRTDIY VTSFGPVSD TEMEYTIDVF FRQSWKDERL RFKGPMQRLP LNNLLASKIW TPDTFFHNGK KSIAHNMTTP NKLLRLEDDG T LLYTMRLT ISAECPMQLE DFPMDAHACP LKFGSYAYPN SEVVYVWTNG STKSVVVAED GSRLNQYHLM GQTVGTENIS TS TGEYTIM TAHFHLKRKI GYFVIQTYLP CIMTVILSQV SFWLNRESVP ARTVFGVTTV LTMTTLSISA RNSLPKVAYA TAM DWFIAV CYAFVFSALI EFATVNYFTK SQPARAAKID KMSRIVFPVL FGTFNLVYWA TYLNREPVIK GAASPK UniProtKB: Gamma-aminobutyric acid receptor subunit alpha-5, Gamma-aminobutyric acid receptor subunit alpha-5 |
-Macromolecule #8: GAMMA-AMINO-BUTANOIC ACID
| Macromolecule | Name: GAMMA-AMINO-BUTANOIC ACID / type: ligand / ID: 8 / Number of copies: 1 / Formula: ABU |
|---|---|
| Molecular weight | Theoretical: 103.12 Da |
| Chemical component information |  ChemComp-ABU: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 200 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average exposure time: 8.0 sec. / Average electron dose: 56.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)