[English] 日本語
 Yorodumi
Yorodumi- EMDB-6552: Cryo-EM structure of the magnesium channel CorA in the magnesium-... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6552 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the magnesium channel CorA in the magnesium-free asymmetric open state I | |||||||||
 Map data Map data | Reconstruction of Thermotoga maritima CorA in the absence of magnesium resulting in at least two conformations; here state I | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | membrane protein / ion channel / magnesium channel / pentameric complex / symmetry vs. asymmetry / conformational change / direct electron detector / K2 / single-particle cryo-electron microscopy | |||||||||
| Function / homology |  Function and homology information Function and homology informationcobalt ion transport / magnesium ion transmembrane transport / cobalt ion transmembrane transporter activity / magnesium ion transmembrane transporter activity / cobalt ion binding / protein homooligomerization / magnesium ion binding / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Thermotoga maritima (bacteria) Thermotoga maritima (bacteria) | |||||||||
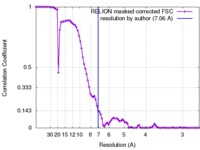
| Method | single particle reconstruction / cryo EM / Resolution: 7.06 Å | |||||||||
 Authors Authors | Matthies D / Dalmas O / Borgnia MJ / Dominik PK / Merk A / Rao P / Reddy BG / Islam S / Bartesaghi A / Perozo E / Subramaniam S | |||||||||
 Citation Citation |  Journal: Cell / Year: 2016 Journal: Cell / Year: 2016Title: Cryo-EM Structures of the Magnesium Channel CorA Reveal Symmetry Break upon Gating. Authors: Doreen Matthies / Olivier Dalmas / Mario J Borgnia / Pawel K Dominik / Alan Merk / Prashant Rao / Bharat G Reddy / Shahidul Islam / Alberto Bartesaghi / Eduardo Perozo / Sriram Subramaniam /  Abstract: CorA, the major Mg(2+) uptake system in prokaryotes, is gated by intracellular Mg(2+) (KD ∼ 1-2 mM). X-ray crystallographic studies of CorA show similar conformations under Mg(2+)-bound and Mg(2+)- ...CorA, the major Mg(2+) uptake system in prokaryotes, is gated by intracellular Mg(2+) (KD ∼ 1-2 mM). X-ray crystallographic studies of CorA show similar conformations under Mg(2+)-bound and Mg(2+)-free conditions, but EPR spectroscopic studies reveal large Mg(2+)-driven quaternary conformational changes. Here, we determined cryo-EM structures of CorA in the Mg(2+)-bound closed conformation and in two open Mg(2+)-free states at resolutions of 3.8, 7.1, and 7.1 Å, respectively. In the absence of bound Mg(2+), four of the five subunits are displaced to variable extents (∼ 10-25 Å) by hinge-like motions as large as ∼ 35° at the stalk helix. The transition between a single 5-fold symmetric closed state and an ensemble of low Mg(2+), open, asymmetric conformational states is, thus, the key structural signature of CorA gating. This mechanism is likely to apply to other structurally similar divalent ion channels. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6552.map.gz emd_6552.map.gz | 59.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6552-v30.xml emd-6552-v30.xml emd-6552.xml emd-6552.xml | 12.5 KB 12.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_6552_fsc.xml emd_6552_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_6552.png emd_6552.png emd_6552_1.png emd_6552_1.png | 167.2 KB 397.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6552 http://ftp.pdbj.org/pub/emdb/structures/EMD-6552 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6552 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6552 | HTTPS FTP |
-Validation report
| Summary document |  emd_6552_validation.pdf.gz emd_6552_validation.pdf.gz | 371.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_6552_full_validation.pdf.gz emd_6552_full_validation.pdf.gz | 371.5 KB | Display | |
| Data in XML |  emd_6552_validation.xml.gz emd_6552_validation.xml.gz | 10.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6552 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6552 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6552 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6552 | HTTPS FTP |
-Related structure data
| Related structure data |  3jcgMC  6551C  6553C  3jcfC  3jchC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6552.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6552.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of Thermotoga maritima CorA in the absence of magnesium resulting in at least two conformations; here state I | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.352 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
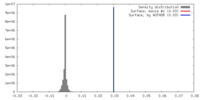
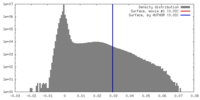
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : CorA from Thermotoga maritima in the absence of magnesium, state I
| Entire | Name: CorA from Thermotoga maritima in the absence of magnesium, state I |
|---|---|
| Components |
|
-Supramolecule #1000: CorA from Thermotoga maritima in the absence of magnesium, state I
| Supramolecule | Name: CorA from Thermotoga maritima in the absence of magnesium, state I type: sample / ID: 1000 / Details: Detergent-solubilized, purified protein / Oligomeric state: One homopentamer of CorA / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 200 KDa |
-Macromolecule #1: CorA
| Macromolecule | Name: CorA / type: protein_or_peptide / ID: 1 / Name.synonym: Magnesium Channel CorA / Number of copies: 5 / Oligomeric state: Pentamer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Thermotoga maritima (bacteria) / Location in cell: Inner Membrane Thermotoga maritima (bacteria) / Location in cell: Inner Membrane |
| Molecular weight | Theoretical: 200 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: Cobalt/magnesium transport protein CorA / InterPro: Magnesium/cobalt transport protein CorA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.7 mg/mL |
|---|---|
| Buffer | pH: 7.3 / Details: 50 mM HEPES, 150 mM NaCl, 1 mM EDTA, 0.5 mM DDM |
| Grid | Details: 300 mesh Cu R1.2/1.3 holey carbon grids from Quantifoil, plasma-cleaned |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 86 % / Chamber temperature: 90 K / Instrument: LEICA EM GP Method: Grids were blotted at 4 degrees Celsius for 7 seconds after a 10-second pre-blotting period, then plunge-frozen in liquid ethane. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 79.6 K / Max: 79.8 K / Average: 79.7 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 105,000 times magnification. Legacy - Electron beam tilt params: 5 |
| Specialist optics | Energy filter - Name: Gatan, Inc. / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 20.0 eV |
| Details | Parallel beam illumination |
| Date | Oct 15, 2014 |
| Image recording | Category: CCD / Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Number real images: 2498 / Average electron dose: 40 e/Å2 Details: Every image is the average of 38 frames recorded by the direct electron detector. The total exposure time was 15.2 seconds, and intermediate frames were recorded every 0.4 seconds. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 105000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.7 µm / Nominal defocus min: 0.89 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder: Liquid nitrogen-cooled / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)