+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of bacteriophage T5 tail tube | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Complex / VIRAL PROTEIN | |||||||||||||||
| Function / homology | virus tail, tube / symbiont genome ejection through host cell envelope, long flexible tail mechanism / Bacterial Ig-like domain (group 2) / Invasin/intimin cell-adhesion fragments / Bacterial Ig-like domain 2 / Bacterial Ig-like domain, group 2 / virus tail / viral release from host cell by cytolysis / Tail tube protein pb6 Function and homology information Function and homology information | |||||||||||||||
| Biological species |  Escherichia phage T5 (virus) Escherichia phage T5 (virus) | |||||||||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||||||||
 Authors Authors | Peng YN / Liu HR | |||||||||||||||
| Funding support |  China, 4 items China, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Int J Mol Sci / Year: 2024 Journal: Int J Mol Sci / Year: 2024Title: Structures of Mature and Urea-Treated Empty Bacteriophage T5: Insights into Siphophage Infection and DNA Ejection. Authors: Yuning Peng / Huanrong Tang / Hao Xiao / Wenyuan Chen / Jingdong Song / Jing Zheng / Hongrong Liu /  Abstract: T5 is a siphophage that has been extensively studied by structural and biochemical methods. However, the complete in situ structures of T5 before and after DNA ejection remain unknown. In this study, ...T5 is a siphophage that has been extensively studied by structural and biochemical methods. However, the complete in situ structures of T5 before and after DNA ejection remain unknown. In this study, we used cryo-electron microscopy (cryo-EM) to determine the structures of mature T5 (a laboratory-adapted, fiberless T5 mutant) and urea-treated empty T5 (lacking the tip complex) at near-atomic resolutions. Atomic models of the head, connector complex, tail tube, and tail tip were built for mature T5, and atomic models of the connector complex, comprising the portal protein pb7, adaptor protein p144, and tail terminator protein p142, were built for urea-treated empty T5. Our findings revealed that the aforementioned proteins did not undergo global conformational changes before and after DNA ejection, indicating that these structural features were conserved among most myophages and siphophages. The present study elucidates the underlying mechanisms of siphophage infection and DNA ejection. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_60712.map.gz emd_60712.map.gz | 27.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-60712-v30.xml emd-60712-v30.xml emd-60712.xml emd-60712.xml | 13.8 KB 13.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_60712.png emd_60712.png | 70.8 KB | ||
| Filedesc metadata |  emd-60712.cif.gz emd-60712.cif.gz | 5 KB | ||
| Others |  emd_60712_half_map_1.map.gz emd_60712_half_map_1.map.gz emd_60712_half_map_2.map.gz emd_60712_half_map_2.map.gz | 22.8 MB 22.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-60712 http://ftp.pdbj.org/pub/emdb/structures/EMD-60712 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60712 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60712 | HTTPS FTP |
-Validation report
| Summary document |  emd_60712_validation.pdf.gz emd_60712_validation.pdf.gz | 877.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_60712_full_validation.pdf.gz emd_60712_full_validation.pdf.gz | 877.3 KB | Display | |
| Data in XML |  emd_60712_validation.xml.gz emd_60712_validation.xml.gz | 10.2 KB | Display | |
| Data in CIF |  emd_60712_validation.cif.gz emd_60712_validation.cif.gz | 11.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60712 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60712 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60712 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60712 | HTTPS FTP |
-Related structure data
| Related structure data |  9inyMC  8zviC  9ilpC  9ilvC 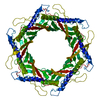 9imhC  9imvC  9iozC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_60712.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_60712.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3 Å | ||||||||||||||||||||||||||||||||||||
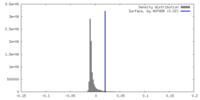
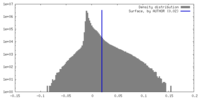
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_60712_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
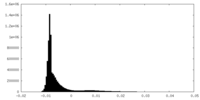
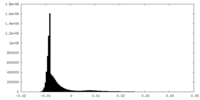
| Density Histograms |
-Half map: #1
| File | emd_60712_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
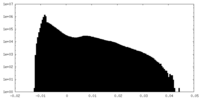
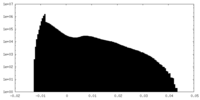
| Density Histograms |
- Sample components
Sample components
-Entire : Escherichia phage T5
| Entire | Name:  Escherichia phage T5 (virus) Escherichia phage T5 (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Escherichia phage T5
| Supramolecule | Name: Escherichia phage T5 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 2695836 / Sci species name: Escherichia phage T5 / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Tail tube protein pb6
| Macromolecule | Name: Tail tube protein pb6 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage T5 (virus) Escherichia phage T5 (virus) |
| Molecular weight | Theoretical: 50.459215 KDa |
| Sequence | String: MSLQLLRNTR IFVSTVKTGH NKTNTQEILV QDDISWGQDS NSTDITVNEA GPRPTRGSKR FNDSLNAAEW SFSTYILPYK DKNTSKQIV PDYMLWHALS SGRAINLEGT TGAHNNATNF MVNFKDNSYH ELAMLHIYIL TDKTWSYIDS CQINQAEVNV D IEDIGRVT ...String: MSLQLLRNTR IFVSTVKTGH NKTNTQEILV QDDISWGQDS NSTDITVNEA GPRPTRGSKR FNDSLNAAEW SFSTYILPYK DKNTSKQIV PDYMLWHALS SGRAINLEGT TGAHNNATNF MVNFKDNSYH ELAMLHIYIL TDKTWSYIDS CQINQAEVNV D IEDIGRVT WSGNGNQLIP LDEQPFDPDQ IGIDDETYMT IQGSYIKNKL TILKIKDMDT NKSYDIPITG GTFTINNNIT YL TPNVMSR VTIPIGSFTG AFELTGSLTA YLNDKSLGSM ELYKDLIKTL KVVNRFEIAL VLGGEYDDER PAAILVAKQA HVN IPTIET DDVLGTSVEF KAIPSDLDAG DEGYLGFSSK YTRTTINNLI VNGDGATDAV TAITVKSAGN VTTLNRSATL QMSV EVTPS SARNKEVTWA ITAGDAATIN ATGLLRADAS KTGAVTVEAT AKDGSGVKGT KVITVTAGG UniProtKB: Tail tube protein pb6 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 32.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 39.0 Å Applied symmetry - Helical parameters - Δ&Phi: 40 ° Applied symmetry - Helical parameters - Axial symmetry: C3 (3 fold cyclic) Resolution.type: BY AUTHOR / Resolution: 3.6 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 199483 |
|---|---|
| Startup model | Type of model: NONE |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)