+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the bacteriophage T5 capsid | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Complex / VIRAL PROTEIN / STRUCTURAL PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationviral scaffold / viral capsid, decoration / T=13 icosahedral viral capsid / symbiont-mediated evasion of host immune response / viral capsid Similarity search - Function | |||||||||||||||
| Biological species |  Escherichia phage T5 (virus) Escherichia phage T5 (virus) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||||||||
 Authors Authors | Peng Y / Liu HR | |||||||||||||||
| Funding support |  China, 4 items China, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Int J Mol Sci / Year: 2024 Journal: Int J Mol Sci / Year: 2024Title: Structures of Mature and Urea-Treated Empty Bacteriophage T5: Insights into Siphophage Infection and DNA Ejection. Authors: Yuning Peng / Huanrong Tang / Hao Xiao / Wenyuan Chen / Jingdong Song / Jing Zheng / Hongrong Liu /  Abstract: T5 is a siphophage that has been extensively studied by structural and biochemical methods. However, the complete in situ structures of T5 before and after DNA ejection remain unknown. In this study, ...T5 is a siphophage that has been extensively studied by structural and biochemical methods. However, the complete in situ structures of T5 before and after DNA ejection remain unknown. In this study, we used cryo-electron microscopy (cryo-EM) to determine the structures of mature T5 (a laboratory-adapted, fiberless T5 mutant) and urea-treated empty T5 (lacking the tip complex) at near-atomic resolutions. Atomic models of the head, connector complex, tail tube, and tail tip were built for mature T5, and atomic models of the connector complex, comprising the portal protein pb7, adaptor protein p144, and tail terminator protein p142, were built for urea-treated empty T5. Our findings revealed that the aforementioned proteins did not undergo global conformational changes before and after DNA ejection, indicating that these structural features were conserved among most myophages and siphophages. The present study elucidates the underlying mechanisms of siphophage infection and DNA ejection. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_60511.map.gz emd_60511.map.gz | 163.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-60511-v30.xml emd-60511-v30.xml emd-60511.xml emd-60511.xml | 15.8 KB 15.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_60511.png emd_60511.png | 129.4 KB | ||
| Filedesc metadata |  emd-60511.cif.gz emd-60511.cif.gz | 5.3 KB | ||
| Others |  emd_60511_half_map_1.map.gz emd_60511_half_map_1.map.gz emd_60511_half_map_2.map.gz emd_60511_half_map_2.map.gz | 161.5 MB 161.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-60511 http://ftp.pdbj.org/pub/emdb/structures/EMD-60511 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60511 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60511 | HTTPS FTP |
-Validation report
| Summary document |  emd_60511_validation.pdf.gz emd_60511_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_60511_full_validation.pdf.gz emd_60511_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_60511_validation.xml.gz emd_60511_validation.xml.gz | 15.3 KB | Display | |
| Data in CIF |  emd_60511_validation.cif.gz emd_60511_validation.cif.gz | 18.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60511 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60511 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60511 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60511 | HTTPS FTP |
-Related structure data
| Related structure data |  8zviMC  9ilpC  9ilvC 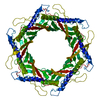 9imhC  9imvC  9inyC  9iozC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_60511.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_60511.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_60511_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_60511_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Escherichia phage T5
| Entire | Name:  Escherichia phage T5 (virus) Escherichia phage T5 (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Escherichia phage T5
| Supramolecule | Name: Escherichia phage T5 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 2695836 / Sci species name: Escherichia phage T5 / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Major capsid protein
| Macromolecule | Name: Major capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage T5 (virus) Escherichia phage T5 (virus) |
| Molecular weight | Theoretical: 50.951684 KDa |
| Sequence | String: MTIDINKLKE ELGLGDLAKS LEGLTAAQKA QEAERMRKEQ EEKELARMND LVSKAVGEDR KRLEEALELV KSLDEKSKKS NELFAQTVE KQQETIVGLQ DEIKSLLTAR EGRSFVGDSV AKALYGTQEN FEDEVEKLVL LSYVMEKGVF ETEHGQRHLK A VNQSSSVE ...String: MTIDINKLKE ELGLGDLAKS LEGLTAAQKA QEAERMRKEQ EEKELARMND LVSKAVGEDR KRLEEALELV KSLDEKSKKS NELFAQTVE KQQETIVGLQ DEIKSLLTAR EGRSFVGDSV AKALYGTQEN FEDEVEKLVL LSYVMEKGVF ETEHGQRHLK A VNQSSSVE VSSESYETIF SQRIIRDLQK ELVVGALFEE LPMSSKILTM LVEPDAGKAT WVAASTYGTD TTTGEEVKGA LK EIHFSTY KLAAKSFITD ETEEDAIFSL LPLLRKRLIE AHAVSIEEAF MTGDGSGKPK GLLTLASEDS AKVVTEAKAD GSV LVTAKT ISKLRRKLGR HGLKLSKLVL IVSMDAYYDL LEDEEWQDVA QVGNDSVKLQ GQVGRIYGLP VVVSEYFPAK ANSA EFAVI VYKDNFVMPR QRAVTVERER QAGKQRDAYY VTQRVNLQRY FANGVVSGTY AAS UniProtKB: Major capsid protein |
-Macromolecule #2: Decoration protein
| Macromolecule | Name: Decoration protein / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage T5 (virus) Escherichia phage T5 (virus) |
| Molecular weight | Theoretical: 17.265447 KDa |
| Sequence | String: MIDYSGLRTI FGEKLPESHI FFATVAAHKY VPSYAFLRRE LGLSSAHTNR KVWKKFVEAY GKAIPPAPPA PPLTLSKDLT ASMSVEEGA ALTLSVTATG GTGPYTYAWT KDGSPIPDAS GATYTKPTAA AEDAGSYKVT VTDSKQVSKD STTCAVTVNP T VPGG UniProtKB: Decoration protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 32.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 100841 |
| Initial angle assignment | Type: COMMON LINE |
| Final angle assignment | Type: COMMON LINE |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)