+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | hAE3NTD2TMD with PT5,CLR, and Y01 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TRANSPORT PROTEIN | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
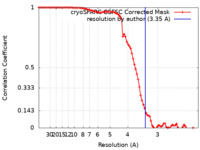
| Method | single particle reconstruction / cryo EM / Resolution: 3.35 Å | |||||||||
 Authors Authors | Jian L / Zhang Q / Yao D / Wang Q / Xia Y / Qin A / Cao Y | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: The structural insight into the functional modulation of human anion exchanger 3. Authors: Liyan Jian / Qing Zhang / Deqiang Yao / Qian Wang / Moxin Chen / Ying Xia / Shaobai Li / Yafeng Shen / Mi Cao / An Qin / Lin Li / Yu Cao /   Abstract: Anion exchanger 3 (AE3) is pivotal in regulating intracellular pH across excitable tissues, yet its structural intricacies and functional dynamics remain underexplored compared to other anion ...Anion exchanger 3 (AE3) is pivotal in regulating intracellular pH across excitable tissues, yet its structural intricacies and functional dynamics remain underexplored compared to other anion exchangers. This study unveils the structural insights into human AE3, including the cryo-electron microscopy structures for AE3 transmembrane domains (TMD) and a chimera combining AE3 N-terminal domain (NTD) with AE2 TMD (hAE32). Our analyzes reveal a substrate binding site, an NTD-TMD interlock mechanism, and a preference for an outward-facing conformation. Unlike AE2, which has more robust acid-loading capabilities, AE3's structure, including a less stable inward-facing conformation due to missing key NTD-TMD interactions, contributes to its moderated pH-modulating activity and increased sensitivity to the inhibitor DIDS. These structural differences underline AE3's distinct functional roles in specific tissues and underscore the complex interplay between structural dynamics and functional specificity within the anion exchanger family, enhancing our understanding of the physiological and pathological roles of the anion exchanger family. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_60225.map.gz emd_60225.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-60225-v30.xml emd-60225-v30.xml emd-60225.xml emd-60225.xml | 15.7 KB 15.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_60225_fsc.xml emd_60225_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_60225.png emd_60225.png | 55.6 KB | ||
| Filedesc metadata |  emd-60225.cif.gz emd-60225.cif.gz | 6.3 KB | ||
| Others |  emd_60225_half_map_1.map.gz emd_60225_half_map_1.map.gz emd_60225_half_map_2.map.gz emd_60225_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-60225 http://ftp.pdbj.org/pub/emdb/structures/EMD-60225 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60225 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60225 | HTTPS FTP |
-Validation report
| Summary document |  emd_60225_validation.pdf.gz emd_60225_validation.pdf.gz | 846.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_60225_full_validation.pdf.gz emd_60225_full_validation.pdf.gz | 846.5 KB | Display | |
| Data in XML |  emd_60225_validation.xml.gz emd_60225_validation.xml.gz | 16.5 KB | Display | |
| Data in CIF |  emd_60225_validation.cif.gz emd_60225_validation.cif.gz | 21.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60225 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60225 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60225 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60225 | HTTPS FTP |
-Related structure data
| Related structure data |  8zleMC  8y85C  8y86C  8y8kC M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_60225.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_60225.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
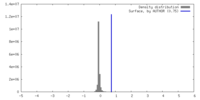
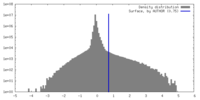
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_60225_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
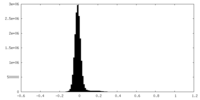
| Density Histograms |
-Half map: #2
| File | emd_60225_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
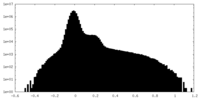
| Density Histograms |
- Sample components
Sample components
-Entire : the NTD of hAE3 with the TMD of hAE2
| Entire | Name: the NTD of hAE3 with the TMD of hAE2 |
|---|---|
| Components |
|
-Supramolecule #1: the NTD of hAE3 with the TMD of hAE2
| Supramolecule | Name: the NTD of hAE3 with the TMD of hAE2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 105 KDa |
-Macromolecule #1: the NTD of hAE3 with the TMD of hAE2
| Macromolecule | Name: the NTD of hAE3 with the TMD of hAE2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 137.4155 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MANGVIPPPG GASPLPQVRV PLEEPPLSPD VEEEDDDLGK TLAVSRFGDL ISKPPAWDPE KPSRSYSERD FEFHRHTSHH THHPLSARL PPPHKLRRLP PTSARHTRRK RKKEKTSAPP SEGTPPIQEE GGAGVDEEEE EEEEEEGESE AEPVEPPHSG T PQKAKFSI ...String: MANGVIPPPG GASPLPQVRV PLEEPPLSPD VEEEDDDLGK TLAVSRFGDL ISKPPAWDPE KPSRSYSERD FEFHRHTSHH THHPLSARL PPPHKLRRLP PTSARHTRRK RKKEKTSAPP SEGTPPIQEE GGAGVDEEEE EEEEEEGESE AEPVEPPHSG T PQKAKFSI GSDEDDSPGL PGRAAVTKPL PSVGPHTDKS PQHSSSSPSP RARASRLAGE KSRPWSPSAS YDLRERLCPG SA LGNPGGP EQQVPTDEAE AQMLGSADLD DMKSHRLEDN PGVRRHLVKK PSRTQGGRGS PSGLAPILRR KKKKKKLDRR PHE VFVELN ELMLDRSQEP HWRETARWIK FEEDVEEETE RWGKPHVASL SFRSLLELRR TIAHGAALLD LEQTTLPGIA HLVV ETMIV SDQIRPEDRA SVLRTLLLKH SHPNDDKDSG FFPRNPSSSS MNSVLGNHHP TPSHGPDGAV PTMADDLGEP APLWP HDPD AKEKPLHMPG GDGHRGKSLK LLEKIPEDAE ATVVLVGCVP FLEQPAAAFV RLNEAVLLES VLEVPVPVRF LFVMLG PSH TSTDYHELGR SIATLMSDKL FHEAAYQADD RQDLLSAISE FLDGSIVIPP SEVEGRDLLR SVAAFQRELL RKRRERE QT KVEMTTRGGY TAPGKELSLE LGGSEATPED DPLRRTGRPF GGLIRDVRRR YPHYLSDFRD ALDPQCLAAV IFIYFAAL S PAITFGGLLG EKTQDLIGVS ELIMSTALQG VVFCLLGAQP LLVIGFSGPL LVFEEAFFSF CSSNHLEYLV GRVWIGFWL VFLALLMVAL EGSFLVRFVS RFTQEIFAFL ISLIFIYETF YKLVKIFQEH PLHGCSASNS SEVDGGENMT WAGARPTLGP GNRSLAGQS GQGKPRGQPN TALLSLVLMA GTFFIAFFLR KFKNSRFFPG RIRRVIGDFG VPIAILIMVL VDYSIEDTYT Q KLSVPSGF SVTAPEKRGW VINPLGEKSP FPVWMMVASL LPAILVFILI FMETQITTLI ISKKERMLQK GSGFHLDLLL IV AMGGICA LFGLPWLAAA TVRSVTHANA LTVMSKAVAP GDKPKIQEVK EQRVTGLLVA LLVGLSIVIG DLLRQIPLAV LFG IFLYMG VTSLNGIQFY ERLHLLLMPP KHHPDVTYVK KVRTLRMHLF TALQLLCLAL LWAVMSTAAS LAFPFILILT VPLR MVVLT RIFTDREMKC LDANEAEPVF DEREGVDEYN EMPMPV |
-Macromolecule #2: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 2 / Number of copies: 2 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #3: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 3 / Number of copies: 2 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Macromolecule #4: [(2R)-1-octadecanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tr...
| Macromolecule | Name: [(2R)-1-octadecanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phospho ryl]oxy-propan-2-yl] (8Z)-icosa-5,8,11,14-tetraenoate type: ligand / ID: 4 / Number of copies: 2 / Formula: PT5 |
|---|---|
| Molecular weight | Theoretical: 1.047088 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)