[English] 日本語
 Yorodumi
Yorodumi- EMDB-5838: Characterization of Human Papilloma Virus Type 11 VLP decorated w... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5838 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Characterization of Human Papilloma Virus Type 11 VLP decorated with antibody fragment H11.B2 B2 | |||||||||
 Map data Map data | Reconstruction of HPV Type 11 VLP + H11.B2 B2 Fab | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Human Papilloma Virus VLP | |||||||||
| Biological species | unidentified (others) /   Human papillomavirus Human papillomavirus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 20.0 Å | |||||||||
 Authors Authors | Zhao Q / Potter CS / Carragher B / Lander G / Sworen J / Towne V / Abraham D / Duncan P / Washabaugh MW / Sitrin RD | |||||||||
 Citation Citation |  Journal: Hum Vaccin Immunother / Year: 2014 Journal: Hum Vaccin Immunother / Year: 2014Title: Characterization of virus-like particles in GARDASIL® by cryo transmission electron microscopy. Authors: Qinjian Zhao / Clinton S Potter / Bridget Carragher / Gabriel Lander / Jaime Sworen / Victoria Towne / Dicky Abraham / Paul Duncan / Michael W Washabaugh / Robert D Sitrin /  Abstract: Cryo-transmission electron microscopy (cryoTEM) is a powerful characterization method for assessing the structural properties of biopharmaceutical nanoparticles, including Virus Like Particle-based ...Cryo-transmission electron microscopy (cryoTEM) is a powerful characterization method for assessing the structural properties of biopharmaceutical nanoparticles, including Virus Like Particle-based vaccines. We demonstrate the method using the Human Papilloma Virus (HPV) VLPs in GARDASIL®. CryoTEM, coupled to automated data collection and analysis, was used to acquire images of the particles in their hydrated state, determine their morphological characteristics, and confirm the integrity of the particles when absorbed to aluminum adjuvant. In addition, we determined the three-dimensional structure of the VLPs, both alone and when interacting with neutralizing antibodies. Two modes of binding of two different neutralizing antibodies were apparent; for HPV type 11 saturated with H11.B2, 72 potential Fab binding sites were observed at the center of each capsomer, whereas for HPV 16 interacting with H16.V5, it appears that 60 pentamers (each neighboring 6 other pentamers) bind five Fabs per pentamer, for the total of 300 potential Fab binding sites per VLP. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5838.map.gz emd_5838.map.gz | 31.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5838-v30.xml emd-5838-v30.xml emd-5838.xml emd-5838.xml | 8.4 KB 8.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5838.png emd_5838.png | 135.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5838 http://ftp.pdbj.org/pub/emdb/structures/EMD-5838 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5838 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5838 | HTTPS FTP |
-Validation report
| Summary document |  emd_5838_validation.pdf.gz emd_5838_validation.pdf.gz | 79 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_5838_full_validation.pdf.gz emd_5838_full_validation.pdf.gz | 78.1 KB | Display | |
| Data in XML |  emd_5838_validation.xml.gz emd_5838_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5838 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5838 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5838 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5838 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_5838.map.gz / Format: CCP4 / Size: 41.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5838.map.gz / Format: CCP4 / Size: 41.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of HPV Type 11 VLP + H11.B2 B2 Fab | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.26 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
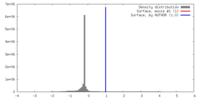
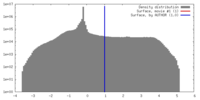
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human Papilloma Virus Type 11 VLP decorated with antibody fragmen...
| Entire | Name: Human Papilloma Virus Type 11 VLP decorated with antibody fragment H11.B2 |
|---|---|
| Components |
|
-Supramolecule #1000: Human Papilloma Virus Type 11 VLP decorated with antibody fragmen...
| Supramolecule | Name: Human Papilloma Virus Type 11 VLP decorated with antibody fragment H11.B2 type: sample / ID: 1000 / Number unique components: 2 |
|---|
-Supramolecule #1: Human papillomavirus
| Supramolecule | Name: Human papillomavirus / type: virus / ID: 1 Details: VLP bound with Type 11 VLP dh antibody fragment H11.B2 NCBI-ID: 10566 / Sci species name: Human papillomavirus / Database: NCBI / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: SEROTYPE / Virus enveloped: No / Virus empty: Yes / Sci species serotype: Type 11 |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) / synonym: VERTEBRATES Homo sapiens (human) / synonym: VERTEBRATES |
-Macromolecule #1: H11.B2 Fab
| Macromolecule | Name: H11.B2 Fab / type: protein_or_peptide / ID: 1 / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism: unidentified (others) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK II |
|---|
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Date | Oct 11, 2006 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) |
| Electron beam | Acceleration voltage: 120 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 20.0 Å / Resolution method: OTHER / Software - Name: EMAN / Number images used: 6209 |
|---|
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)