[English] 日本語
 Yorodumi
Yorodumi- EMDB-5457: Cryo-electron tomography of native, trimeric HIV-1 BaL Env in com... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5457 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-electron tomography of native, trimeric HIV-1 BaL Env in complex with VRC01 IgG | |||||||||
 Map data Map data | Molecular structure of native HIV-1 BaL Env trimer in complex with VRC01 IgG | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HIV / Env / VRC01 / neutralizing antibody | |||||||||
| Biological species |   Human immunodeficiency virus 1 / unidentified (others) Human immunodeficiency virus 1 / unidentified (others) | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 24.0 Å | |||||||||
 Authors Authors | Tran EEH / Borgnia MJ / Kuybeda O / Schauder DM / Bartesaghi A / Frank GA / Sapiro G / Milne JLS / Subramaniam S | |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2012 Journal: PLoS Pathog / Year: 2012Title: Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. Authors: Erin E H Tran / Mario J Borgnia / Oleg Kuybeda / David M Schauder / Alberto Bartesaghi / Gabriel A Frank / Guillermo Sapiro / Jacqueline L S Milne / Sriram Subramaniam /  Abstract: HIV-1 infection begins with the binding of trimeric viral envelope glycoproteins (Env) to CD4 and a co-receptor on target T-cells. Understanding how these ligands influence the structure of Env is of ...HIV-1 infection begins with the binding of trimeric viral envelope glycoproteins (Env) to CD4 and a co-receptor on target T-cells. Understanding how these ligands influence the structure of Env is of fundamental interest for HIV vaccine development. Using cryo-electron microscopy, we describe the contrasting structural outcomes of trimeric Env binding to soluble CD4, to the broadly neutralizing, CD4-binding site antibodies VRC01, VRC03 and b12, or to the monoclonal antibody 17b, a co-receptor mimic. Binding of trimeric HIV-1 BaL Env to either soluble CD4 or 17b alone, is sufficient to trigger formation of the open quaternary conformation of Env. In contrast, VRC01 locks Env in the closed state, while b12 binding requires a partial opening in the quaternary structure of trimeric Env. Our results show that, despite general similarities in regions of the HIV-1 gp120 polypeptide that contact CD4, VRC01, VRC03 and b12, there are important differences in quaternary structures of the complexes these ligands form on native trimeric Env, and potentially explain differences in the neutralizing breadth and potency of antibodies with similar specificities. From cryo-electron microscopic analysis at ∼9 Å resolution of a cleaved, soluble version of trimeric Env, we show that a structural signature of the open Env conformation is a three-helix motif composed of α-helical segments derived from highly conserved, non-glycosylated N-terminal regions of the gp41 trimer. The three N-terminal gp41 helices in this novel, activated Env conformation are held apart by their interactions with the rest of Env, and are less compactly packed than in the post-fusion, six-helix bundle state. These findings suggest a new structural template for designing immunogens that can elicit antibodies targeting HIV at a vulnerable, pre-entry stage. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5457.map.gz emd_5457.map.gz | 741.6 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5457-v30.xml emd-5457-v30.xml emd-5457.xml emd-5457.xml | 11.2 KB 11.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5457.png emd_5457.png | 1.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5457 http://ftp.pdbj.org/pub/emdb/structures/EMD-5457 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5457 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5457 | HTTPS FTP |
-Validation report
| Summary document |  emd_5457_validation.pdf.gz emd_5457_validation.pdf.gz | 78.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_5457_full_validation.pdf.gz emd_5457_full_validation.pdf.gz | 77.4 KB | Display | |
| Data in XML |  emd_5457_validation.xml.gz emd_5457_validation.xml.gz | 495 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5457 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5457 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5457 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5457 | HTTPS FTP |
-Related structure data
| Related structure data |  5455C  5456C  5458C 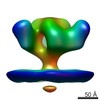 5459C  5460C 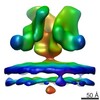 5461C  5462C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5457.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5457.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Molecular structure of native HIV-1 BaL Env trimer in complex with VRC01 IgG | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Molecular structure of native HIV-1 BaL Env trimer in complex wit...
| Entire | Name: Molecular structure of native HIV-1 BaL Env trimer in complex with VRC01 IgG |
|---|---|
| Components |
|
-Supramolecule #1000: Molecular structure of native HIV-1 BaL Env trimer in complex wit...
| Supramolecule | Name: Molecular structure of native HIV-1 BaL Env trimer in complex with VRC01 IgG type: sample / ID: 1000 / Oligomeric state: trimer / Number unique components: 2 |
|---|
-Macromolecule #1: Envelope glycoprotein
| Macromolecule | Name: Envelope glycoprotein / type: protein_or_peptide / ID: 1 / Name.synonym: Env Details: Envelope glycoproteins present on the surface of intact virions. Number of copies: 3 / Oligomeric state: trimer / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 / Strain: BaL / synonym: HIV-1 Human immunodeficiency virus 1 / Strain: BaL / synonym: HIV-1 |
-Macromolecule #2: VRC01 IgG
| Macromolecule | Name: VRC01 IgG / type: protein_or_peptide / ID: 2 / Details: VRC01 neutralizing antibody / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism: unidentified (others) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: TNE Buffer (10 mM Tris, 150 mM NaCl, 1 mM EDTA) |
|---|---|
| Grid | Details: 200 mesh Quantifoil Multi A |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 77 K / Instrument: FEI VITROBOT MARK III Method: blot for 6 seconds, at 25 degrees C, 100 percent humidity, blot offset of -2, plunge into an ethane slurry cooled by liquid nitrogen |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Temperature | Average: 81 K |
| Specialist optics | Energy filter - Name: GATAN GIF / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 20.0 eV |
| Date | Jun 7, 2010 |
| Image recording | Category: CCD / Film or detector model: GENERIC GATAN / Average electron dose: 150 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 2.5 µm / Nominal magnification: 34000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN / Tilt series - Axis1 - Min angle: -60 ° / Tilt series - Axis1 - Max angle: 60 ° |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | Average number of tilts used in the 3D reconstructions: 61. Average tomographic tilt angle increment: 2. |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 24.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name:  IMOD / Details: Resolution for FSC at 0.5 cut-off is 28 Angstrom IMOD / Details: Resolution for FSC at 0.5 cut-off is 28 Angstrom |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - #0 - Chain ID: G / Chain - #1 - Chain ID: H / Chain - #2 - Chain ID: L |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | Protocol: Rigid body. Automated fitting procedures |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller


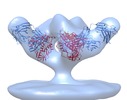

 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)