+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5116 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 30S subunit of ribosomal protein S1 | |||||||||
 Map data Map data | surface view of S1 protein | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | protein S1 / EM density / spider / ribosome / single particle reconstruction | |||||||||
| Biological species | unidentified (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 11.5 Å | |||||||||
 Authors Authors | Sengupta J / Agrawal RK / Frank J | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2001 Journal: Proc Natl Acad Sci U S A / Year: 2001Title: Visualization of protein S1 within the 30S ribosomal subunit and its interaction with messenger RNA. Authors: J Sengupta / R K Agrawal / J Frank /  Abstract: S1 is the largest ribosomal protein, present in the small subunit of the bacterial ribosome. It has a pivotal role in stabilizing the mRNA on the ribosome. Thus far, S1 has eluded structural ...S1 is the largest ribosomal protein, present in the small subunit of the bacterial ribosome. It has a pivotal role in stabilizing the mRNA on the ribosome. Thus far, S1 has eluded structural determination. We have identified the S1 protein mass in the cryo-electron microscopic map of the Escherichia coli ribosome by comparing the map with a recent x-ray crystallographic structure of the 30S subunit, which lacks S1. According to our finding, S1 is located at the junction of head, platform, and main body of the 30S subunit, thus explaining all existing biochemical and crosslinking data. Protein S1 as identified in our map has a complex, elongated shape with two holes in its central portion. The N-terminal domain, forming one of the extensions, penetrates into the head of the 30S subunit. Evidence for direct interaction of S1 with 11 nucleotides of the mRNA, immediately upstream of the Shine-Dalgarno sequence, explains the protein's role in the recognition of the 5' region of mRNA. #1:  Journal: CELL (CAMBRIDGE,MASS.) / Year: 2000 Journal: CELL (CAMBRIDGE,MASS.) / Year: 2000Title: Solution structure of the E. coli 70S ribosome at 11.5 angstrom resolution Authors: Gabashvili IS / Agrawal RK / Spahn CMT / Grassucci RA / Svergun DI / Frank J / Penczek P | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5116.map.gz emd_5116.map.gz | 7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5116-v30.xml emd-5116-v30.xml emd-5116.xml emd-5116.xml | 7.7 KB 7.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5116_1.png emd_5116_1.png | 101.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5116 http://ftp.pdbj.org/pub/emdb/structures/EMD-5116 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5116 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5116 | HTTPS FTP |
-Validation report
| Summary document |  emd_5116_validation.pdf.gz emd_5116_validation.pdf.gz | 77.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_5116_full_validation.pdf.gz emd_5116_full_validation.pdf.gz | 76.5 KB | Display | |
| Data in XML |  emd_5116_validation.xml.gz emd_5116_validation.xml.gz | 493 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5116 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5116 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5116 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5116 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_5116.map.gz / Format: CCP4 / Size: 7.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5116.map.gz / Format: CCP4 / Size: 7.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | surface view of S1 protein | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.93 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
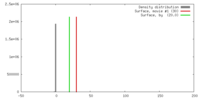
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Protein S1
| Entire | Name: Protein S1 |
|---|---|
| Components |
|
-Supramolecule #1000: Protein S1
| Supramolecule | Name: Protein S1 / type: sample / ID: 1000 Details: see additional reference by Gabashvili I S, Agrawal R K, Spahn C M T, Grassucci R,Svergun D I,and Frank J, 7. Penczek P (2000) Cell 100,537-549 Number unique components: 1 |
|---|
-Macromolecule #1: Protein S1
| Macromolecule | Name: Protein S1 / type: protein_or_peptide / ID: 1 / Name.synonym: Protein S1 / Number of copies: 1 / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism: unidentified (others) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Vitrification | Cryogen name: ETHANE / Instrument: OTHER |
|---|
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 20 |
|---|---|
| Date | Jul 1, 1999 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder: Side entry liquid nitrogen-cooled cryo specimen holder. Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| CTF correction | Details: see the additional reference |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 11.5 Å / Resolution method: OTHER / Software - Name: spider / Details: see the additional reference |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)