+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4567 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Viral pore protein | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | portal / translocase / motor / pore / bacteriophage / thermophage / caudovirales / siphoviridae / virus / VIRAL PROTEIN | |||||||||
| Function / homology | viral capsid / Portal protein Function and homology information Function and homology information | |||||||||
| Biological species |   Thermus phage P2345 (virus) / Thermus phage P2345 (virus) /  Thermus virus P23-45 Thermus virus P23-45 | |||||||||
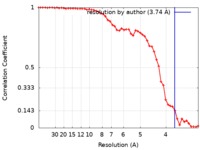
| Method | single particle reconstruction / cryo EM / Resolution: 3.74 Å | |||||||||
 Authors Authors | Bayfield OW / Antson AA / Winkler DC / Hesketh EL / Chechik M / Cheng N / Dykeman EC / Minakhin L / Ranson NA / Severinov K / Steven AC | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2020 Journal: Elife / Year: 2020Title: Cryo-EM structure in situ reveals a molecular switch that safeguards virus against genome loss. Authors: Oliver W Bayfield / Alasdair C Steven / Alfred A Antson /   Abstract: The portal protein is a key component of many double-stranded DNA viruses, governing capsid assembly and genome packaging. Twelve subunits of the portal protein define a tunnel, through which DNA is ...The portal protein is a key component of many double-stranded DNA viruses, governing capsid assembly and genome packaging. Twelve subunits of the portal protein define a tunnel, through which DNA is translocated into the capsid. It is unknown how the portal protein functions as a gatekeeper, preventing DNA slippage, whilst allowing its passage into the capsid, and how these processes are controlled. A cryo-EM structure of the portal protein of thermostable virus P23-45, determined in situ in its procapsid-bound state, indicates a mechanism that naturally safeguards the virus against genome loss. This occurs via an inversion of the conformation of the loops that define the constriction in the central tunnel, accompanied by a hydrophilic-hydrophobic switch. The structure also shows how translocation of DNA into the capsid could be modulated by a changing mode of protein-protein interactions between portal and capsid, across a symmetry-mismatched interface. | |||||||||
| History |
|
- Structure visualization
Structure visualization
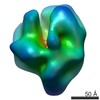
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4567.map.gz emd_4567.map.gz | 9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4567-v30.xml emd-4567-v30.xml emd-4567.xml emd-4567.xml | 16.3 KB 16.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4567_fsc.xml emd_4567_fsc.xml | 5 KB | Display |  FSC data file FSC data file |
| Images |  emd_4567.png emd_4567.png | 183.6 KB | ||
| Masks |  emd_4567_msk_1.map emd_4567_msk_1.map | 10 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-4567.cif.gz emd-4567.cif.gz | 5.8 KB | ||
| Others |  emd_4567_half_map_1.map.gz emd_4567_half_map_1.map.gz emd_4567_half_map_2.map.gz emd_4567_half_map_2.map.gz | 5.8 MB 5.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4567 http://ftp.pdbj.org/pub/emdb/structures/EMD-4567 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4567 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4567 | HTTPS FTP |
-Validation report
| Summary document |  emd_4567_validation.pdf.gz emd_4567_validation.pdf.gz | 436.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_4567_full_validation.pdf.gz emd_4567_full_validation.pdf.gz | 435.7 KB | Display | |
| Data in XML |  emd_4567_validation.xml.gz emd_4567_validation.xml.gz | 10.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4567 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4567 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4567 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4567 | HTTPS FTP |
-Related structure data
| Related structure data |  6qjtMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4567.map.gz / Format: CCP4 / Size: 10 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4567.map.gz / Format: CCP4 / Size: 10 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.5975 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
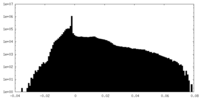
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_4567_msk_1.map emd_4567_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
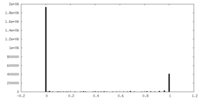
| Density Histograms |
-Half map: #1
| File | emd_4567_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
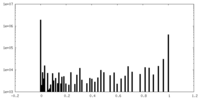
| Density Histograms |
-Half map: #2
| File | emd_4567_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
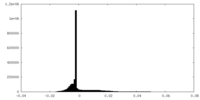
| Density Histograms |
- Sample components
Sample components
-Entire : Thermus phage P2345
| Entire | Name:   Thermus phage P2345 (virus) Thermus phage P2345 (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Thermus phage P2345
| Supramolecule | Name: Thermus phage P2345 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Thermus phage P2345 (virus) Thermus phage P2345 (virus) |
| Molecular weight | Theoretical: 596 KDa |
-Macromolecule #1: Portal protein
| Macromolecule | Name: Portal protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Thermus virus P23-45 Thermus virus P23-45 |
| Molecular weight | Theoretical: 49.714141 KDa |
| Sequence | String: MAKRGRKPKE LVPGPGSIDP SDVPKLEGAS VPVMSTSYDV VVDREFDELL QGKDGLLVYH KMLSDGTVKN ALNYIFGRIR SAKWYVEPA STDPEDIAIA AFIHAQLGID DASVGKYPFG RLFAIYENAY IYGMAAGEIV LTLGADGKLI LDKIVPIHPF N IDEVLYDE ...String: MAKRGRKPKE LVPGPGSIDP SDVPKLEGAS VPVMSTSYDV VVDREFDELL QGKDGLLVYH KMLSDGTVKN ALNYIFGRIR SAKWYVEPA STDPEDIAIA AFIHAQLGID DASVGKYPFG RLFAIYENAY IYGMAAGEIV LTLGADGKLI LDKIVPIHPF N IDEVLYDE EGGPKALKLS GEVKGGSQFV NGLEIPIWKT VVFLHNDDGS FTGQSALRAA VPHWLAKRAL ILLINHGLER FM IGVPTLT IPKSVRQGTK QWEAAKEIVK NFVQKPRHGI ILPDDWKFDT VDLKSAMPDA IPYLTYHDAG IARALGIDFN TVQ LNMGVQ AVNIGEFVSL TQQTIISLQR EFASAVNLYL IPKLVLPNWP GATRFPRLTF EMEERNDFSA AANLMGMLIN AVKD SEDIP TELKALIDAL PSKMRRALGV VDEVREAVRQ PADSRYLYTR RRR UniProtKB: Portal protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Average exposure time: 2.0 sec. / Average electron dose: 99.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 75000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-6qjt: |
 Movie
Movie Controller
Controller










 Z
Z Y
Y X
X