[English] 日本語
 Yorodumi
Yorodumi- EMDB-44482: Cryo-EM structure of the HIV-1 JR-FL IDL Env trimer in complex wi... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the HIV-1 JR-FL IDL Env trimer in complex with PGT122 Fab | |||||||||
 Map data Map data | sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CD4 / HIV-1 / SOSIP / Vaccine / gp120 / gp41 / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / virus-mediated perturbation of host defense response / fusion of virus membrane with host endosome membrane / viral envelope ...positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / virus-mediated perturbation of host defense response / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / structural molecule activity / virion membrane / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | |||||||||
 Authors Authors | Gorman J / Kwong PD | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2024 Journal: Cell Rep / Year: 2024Title: Design of soluble HIV-1 envelope trimers free of covalent gp120-gp41 bonds with prevalent native-like conformation. Authors: Peng Zhang / Jason Gorman / Yaroslav Tsybovsky / Maolin Lu / Qingbo Liu / Vinay Gopan / Mamta Singh / Yin Lin / Huiyi Miao / Yuna Seo / Alice Kwon / Adam S Olia / Gwo-Yu Chuang / Hui Geng / ...Authors: Peng Zhang / Jason Gorman / Yaroslav Tsybovsky / Maolin Lu / Qingbo Liu / Vinay Gopan / Mamta Singh / Yin Lin / Huiyi Miao / Yuna Seo / Alice Kwon / Adam S Olia / Gwo-Yu Chuang / Hui Geng / Yen-Ting Lai / Tongqing Zhou / John R Mascola / Walther Mothes / Peter D Kwong / Paolo Lusso /   Abstract: Soluble HIV-1 envelope (Env) trimers may serve as effective vaccine immunogens. The widely utilized SOSIP trimers have been paramount for structural studies, but the disulfide bond they feature ...Soluble HIV-1 envelope (Env) trimers may serve as effective vaccine immunogens. The widely utilized SOSIP trimers have been paramount for structural studies, but the disulfide bond they feature between gp120 and gp41 constrains intersubunit mobility and may alter antigenicity. Here, we report an alternative strategy to generate stabilized soluble Env trimers free of covalent gp120-gp41 bonds. Stabilization was achieved by introducing an intrasubunit disulfide bond between the inner and outer domains of gp120, defined as interdomain lock (IDL). Correctly folded IDL trimers displaying a native-like antigenic profile were produced for HIV-1 Envs of different clades. Importantly, the IDL design abrogated CD4 binding while not affecting recognition by potent neutralizing antibodies to the CD4-binding site. By cryoelectron microscopy, IDL trimers were shown to adopt a closed prefusion configuration, while single-molecule fluorescence resonance energy transfer documented a high prevalence of native-like conformation. Thus, IDL trimers may be promising candidates as vaccine immunogens. | |||||||||
| History |
|
- Structure visualization
Structure visualization
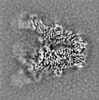
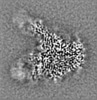
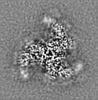
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_44482.map.gz emd_44482.map.gz | 117.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-44482-v30.xml emd-44482-v30.xml emd-44482.xml emd-44482.xml | 25.6 KB 25.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_44482.png emd_44482.png | 79.5 KB | ||
| Masks |  emd_44482_msk_1.map emd_44482_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-44482.cif.gz emd-44482.cif.gz | 7.1 KB | ||
| Others |  emd_44482_additional_1.map.gz emd_44482_additional_1.map.gz emd_44482_additional_2.map.gz emd_44482_additional_2.map.gz emd_44482_half_map_1.map.gz emd_44482_half_map_1.map.gz emd_44482_half_map_2.map.gz emd_44482_half_map_2.map.gz | 29.7 MB 25.2 MB 115.9 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-44482 http://ftp.pdbj.org/pub/emdb/structures/EMD-44482 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44482 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44482 | HTTPS FTP |
-Validation report
| Summary document |  emd_44482_validation.pdf.gz emd_44482_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_44482_full_validation.pdf.gz emd_44482_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_44482_validation.xml.gz emd_44482_validation.xml.gz | 13.9 KB | Display | |
| Data in CIF |  emd_44482_validation.cif.gz emd_44482_validation.cif.gz | 16.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44482 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44482 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44482 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44482 | HTTPS FTP |
-Related structure data
| Related structure data |  9berMC  9bewC  9bf6C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_44482.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_44482.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0733 Å | ||||||||||||||||||||||||||||||||||||
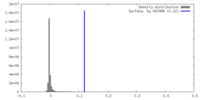
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
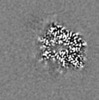
-Mask #1
| File |  emd_44482_msk_1.map emd_44482_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
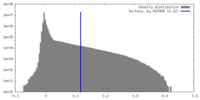
| Density Histograms |
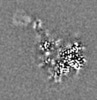
-Additional map: unsharpened map
| File | emd_44482_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
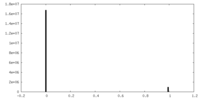
| Density Histograms |
-Additional map: resolve map
| File | emd_44482_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | resolve map | ||||||||||||
| Projections & Slices |
| ||||||||||||
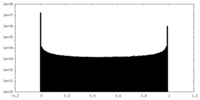
| Density Histograms |
-Half map: half map B
| File | emd_44482_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map A
| File | emd_44482_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM Structure of the HIV-1 JR-FL IDL Env trimer in complex wi...
| Entire | Name: Cryo-EM Structure of the HIV-1 JR-FL IDL Env trimer in complex with PGT122 Fab |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM Structure of the HIV-1 JR-FL IDL Env trimer in complex wi...
| Supramolecule | Name: Cryo-EM Structure of the HIV-1 JR-FL IDL Env trimer in complex with PGT122 Fab type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Macromolecule #1: Envelope glycoprotein gp120
| Macromolecule | Name: Envelope glycoprotein gp120 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 53.686918 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: VEKLWVTVYY GVPVWKEATT TLFCASDAKA YDTEVHNVWA THACVPTDPN PQEVVLENVT EHFNMWKNNM VEQMQEDIIS LWCQSLKPC VKLTPLCVTL NCKDVNATNT TNDSEGTMER GEIKNCSFNI TTSIRDKVQK EYALFYKLDV VPIDNNNTSY R LISCNTSV ...String: VEKLWVTVYY GVPVWKEATT TLFCASDAKA YDTEVHNVWA THACVPTDPN PQEVVLENVT EHFNMWKNNM VEQMQEDIIS LWCQSLKPC VKLTPLCVTL NCKDVNATNT TNDSEGTMER GEIKNCSFNI TTSIRDKVQK EYALFYKLDV VPIDNNNTSY R LISCNTSV ITQACPKISF EPIPIHYCAP AGFAILKCND KTFNGTGPCK NVSTVQCTHG IRPVVSTQLL LNGSLAEEEV VI RSDNFTN NAKTIIVQLK ESVEINCTRP NNNTRKSIHI GPGRAFYTTG EIIGDIRQAH CNISRAKWND TLKQIVIKLR EQF ENKTIV FNHSSGGDPE IVMHSFNCGG EFFYCNSTQL FNSTWNNNTE GSNNTEGNTI TLPCRIKQII NMWQEVGGCG AMYA PPIRG QIRCSSNITG LLLTRDGGIN ENGTEIFRPG GGDMRDNWRS ELYKYKVVKI EPLGVAPTKA KRRVVGRRRR RR UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #2: Envelope glycoprotein gp41
| Macromolecule | Name: Envelope glycoprotein gp41 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 17.227549 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVGIGAVFLG FLGAAGSTMG AASMTLTVQA RLLLSGIVQQ QNNLLRAPEA QQRMLQLTVW GIKQLQARVL AVERYLGDQQ LLGIWGCSG KLICTTAVPW NASWSNKSLD RIWNNMTWME WEREIDNYTS EIYTLIEESQ NQQEKNEQEL LELD UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #3: PGT122 heavy chain
| Macromolecule | Name: PGT122 heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 14.838731 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVHLQESGPG LVKPSETLSL TCNVSGTLVR DNYWSWIRQP LGKQPEWIGY VHDSGDTNYN PSLKSRVHLS LDKSKNLVSL RLTGVTAAD SAIYYCATTK HGRRIYGVVA FKEWFTYFYM DVWGKGTSVT VSS |
-Macromolecule #4: PGT122 light chain
| Macromolecule | Name: PGT122 light chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.423534 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: TFVSVAPGQT ARITCGEESL GSRSVIWYQQ RPGQAPSLII YNNNDRPSGI PDRFSGSPGS TFGTTATLTI TSVEAGDEAD YYCHIWDSR RPTNWVFGEG TTLIVL |
-Macromolecule #8: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 8 / Number of copies: 45 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #9: SULFATE ION
| Macromolecule | Name: SULFATE ION / type: ligand / ID: 9 / Number of copies: 1 / Formula: SO4 |
|---|---|
| Molecular weight | Theoretical: 96.063 Da |
| Chemical component information |  ChemComp-SO4: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: PBS |
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Average exposure time: 10.0 sec. / Average electron dose: 70.73 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.0 µm / Nominal defocus min: 0.4 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)