[English] 日本語
 Yorodumi
Yorodumi- EMDB-43670: Soluble ectodomain of human cytomegalovirus (HCMV) glycoprotein B... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Soluble ectodomain of human cytomegalovirus (HCMV) glycoprotein B (gB) stabilized in a prefusion-like conformation in complex with 1G2 and 7H3, global refinement | |||||||||
 Map data Map data | sharpened | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | orthoherpesvirus / betaherpesvirus / cytomegalovirus / human betaherpesvirus 5 / human cytomegalovirus / HCMV / glycoprotein B / gB / HCMV gB / prefusion-stabilized / disulfide / viral protein / 7H3 / 1G2 | |||||||||
| Biological species |   Human betaherpesvirus 5 / Human betaherpesvirus 5 /  Homo sapiens (human) Homo sapiens (human) | |||||||||
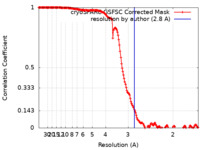
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Sponholtz MR / Byrne PO / McLellan JS | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2024 Journal: Proc Natl Acad Sci U S A / Year: 2024Title: Structure-based design of a soluble human cytomegalovirus glycoprotein B antigen stabilized in a prefusion-like conformation. Authors: Madeline R Sponholtz / Patrick O Byrne / Alison G Lee / Ajit R Ramamohan / Jory A Goldsmith / Ryan S McCool / Ling Zhou / Nicole V Johnson / Ching-Lin Hsieh / Megan Connors / Krithika P ...Authors: Madeline R Sponholtz / Patrick O Byrne / Alison G Lee / Ajit R Ramamohan / Jory A Goldsmith / Ryan S McCool / Ling Zhou / Nicole V Johnson / Ching-Lin Hsieh / Megan Connors / Krithika P Karthigeyan / Chelsea M Crooks / Adelaide S Fuller / John D Campbell / Sallie R Permar / Jennifer A Maynard / Dong Yu / Matthew J Bottomley / Jason S McLellan /  Abstract: Human cytomegalovirus (HCMV) glycoprotein B (gB) is a class III membrane fusion protein required for viral entry. HCMV vaccine candidates containing gB have demonstrated moderate clinical efficacy, ...Human cytomegalovirus (HCMV) glycoprotein B (gB) is a class III membrane fusion protein required for viral entry. HCMV vaccine candidates containing gB have demonstrated moderate clinical efficacy, but no HCMV vaccine has been approved. Here, we used structure-based design to identify and characterize amino acid substitutions that stabilize gB in its metastable prefusion conformation. One variant containing two engineered interprotomer disulfide bonds and two cavity-filling substitutions (gB-C7), displayed increased expression and thermostability. A 2.8 Å resolution cryoelectron microscopy structure shows that gB-C7 adopts a prefusion-like conformation, revealing additional structural elements at the membrane-distal apex. Unlike previous observations for several class I viral fusion proteins, mice immunized with postfusion or prefusion-stabilized forms of soluble gB protein displayed similar neutralizing antibody titers, here specifically against an HCMV laboratory strain on fibroblasts. Collectively, these results identify initial strategies to stabilize class III viral fusion proteins and provide tools to probe gB-directed antibody responses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43670.map.gz emd_43670.map.gz | 398.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43670-v30.xml emd-43670-v30.xml emd-43670.xml emd-43670.xml | 19.5 KB 19.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_43670_fsc.xml emd_43670_fsc.xml | 15.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_43670.png emd_43670.png | 45.9 KB | ||
| Masks |  emd_43670_msk_1.map emd_43670_msk_1.map | 421.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-43670.cif.gz emd-43670.cif.gz | 4.9 KB | ||
| Others |  emd_43670_additional_1.map.gz emd_43670_additional_1.map.gz emd_43670_half_map_1.map.gz emd_43670_half_map_1.map.gz emd_43670_half_map_2.map.gz emd_43670_half_map_2.map.gz | 210.4 MB 391.5 MB 391.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43670 http://ftp.pdbj.org/pub/emdb/structures/EMD-43670 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43670 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43670 | HTTPS FTP |
-Validation report
| Summary document |  emd_43670_validation.pdf.gz emd_43670_validation.pdf.gz | 925.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_43670_full_validation.pdf.gz emd_43670_full_validation.pdf.gz | 925.2 KB | Display | |
| Data in XML |  emd_43670_validation.xml.gz emd_43670_validation.xml.gz | 23.8 KB | Display | |
| Data in CIF |  emd_43670_validation.cif.gz emd_43670_validation.cif.gz | 30.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43670 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43670 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43670 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43670 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_43670.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43670.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8332 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_43670_msk_1.map emd_43670_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: unsharpened
| File | emd_43670_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half B
| File | emd_43670_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half A
| File | emd_43670_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Soluble ectodomain of human cytomegalovirus (HCMV) glycoprotein B...
| Entire | Name: Soluble ectodomain of human cytomegalovirus (HCMV) glycoprotein B (gB) stabilized in a prefusion-like conformation in complex with 1G2 and 7H3 |
|---|---|
| Components |
|
-Supramolecule #1: Soluble ectodomain of human cytomegalovirus (HCMV) glycoprotein B...
| Supramolecule | Name: Soluble ectodomain of human cytomegalovirus (HCMV) glycoprotein B (gB) stabilized in a prefusion-like conformation in complex with 1G2 and 7H3 type: complex / ID: 1 / Parent: 0 |
|---|
-Supramolecule #2: human cytomegalovirus (HCMV) glycoprotein B (gB) ectodomain
| Supramolecule | Name: human cytomegalovirus (HCMV) glycoprotein B (gB) ectodomain type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:   Human betaherpesvirus 5 Human betaherpesvirus 5 |
-Supramolecule #3: Fab 1G2
| Supramolecule | Name: Fab 1G2 / type: complex / ID: 3 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #4: Fab 7H3
| Supramolecule | Name: Fab 7H3 / type: complex / ID: 4 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: 2 mM Tris pH 8, 200 mM NaCl, 0.02% (w/v) sodium azide, 3% (v/v) glycerol, 0.12% (w/v) CHAPS, 0.01% (w/v) amphipol A8-35 | |||||||||||||||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV | |||||||||||||||||||||
| Details | Soluble ectodomain of human cytomegalovirus (HCMV) glycoprotein B (gB) in the stabilized in a prefusion-like conformation, 1G2 Fab, and 7H3 Fab purified separately, mixed, and incubated for 30 minutes on ice prior to grid preparation |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 12524 / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)