[English] 日本語
 Yorodumi
Yorodumi- EMDB-43672: Soluble ectodomain of human cytomegalovirus (HCMV) glycoprotein B... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Soluble ectodomain of human cytomegalovirus (HCMV) glycoprotein B (gB) stabilized in a prefusion-like conformation in complex with 1G2 and 7H3, composite map (global and local) and model | |||||||||
 Map data Map data | Composite Map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | orthoherpesvirus / betaherpesvirus / cytomegalovirus / human betaherpesvirus 5 / human cytomegalovirus / HCMV / glycoprotein B / gB / HCMV gB / prefusion / prefusion-stabilized / disulfide / VIRAL PROTEIN / 7H2 / 1G2 / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell Golgi membrane / host cell endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / virion membrane / membrane Similarity search - Function | |||||||||
| Biological species |   Human betaherpesvirus 5 / Human betaherpesvirus 5 /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Sponholtz MR / Byrne PO / McLellan JS | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2024 Journal: Proc Natl Acad Sci U S A / Year: 2024Title: Structure-based design of a soluble human cytomegalovirus glycoprotein B antigen stabilized in a prefusion-like conformation. Authors: Madeline R Sponholtz / Patrick O Byrne / Alison G Lee / Ajit R Ramamohan / Jory A Goldsmith / Ryan S McCool / Ling Zhou / Nicole V Johnson / Ching-Lin Hsieh / Megan Connors / Krithika P ...Authors: Madeline R Sponholtz / Patrick O Byrne / Alison G Lee / Ajit R Ramamohan / Jory A Goldsmith / Ryan S McCool / Ling Zhou / Nicole V Johnson / Ching-Lin Hsieh / Megan Connors / Krithika P Karthigeyan / Chelsea M Crooks / Adelaide S Fuller / John D Campbell / Sallie R Permar / Jennifer A Maynard / Dong Yu / Matthew J Bottomley / Jason S McLellan /  Abstract: Human cytomegalovirus (HCMV) glycoprotein B (gB) is a class III membrane fusion protein required for viral entry. HCMV vaccine candidates containing gB have demonstrated moderate clinical efficacy, ...Human cytomegalovirus (HCMV) glycoprotein B (gB) is a class III membrane fusion protein required for viral entry. HCMV vaccine candidates containing gB have demonstrated moderate clinical efficacy, but no HCMV vaccine has been approved. Here, we used structure-based design to identify and characterize amino acid substitutions that stabilize gB in its metastable prefusion conformation. One variant containing two engineered interprotomer disulfide bonds and two cavity-filling substitutions (gB-C7), displayed increased expression and thermostability. A 2.8 Å resolution cryoelectron microscopy structure shows that gB-C7 adopts a prefusion-like conformation, revealing additional structural elements at the membrane-distal apex. Unlike previous observations for several class I viral fusion proteins, mice immunized with postfusion or prefusion-stabilized forms of soluble gB protein displayed similar neutralizing antibody titers, here specifically against an HCMV laboratory strain on fibroblasts. Collectively, these results identify initial strategies to stabilize class III viral fusion proteins and provide tools to probe gB-directed antibody responses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43672.map.gz emd_43672.map.gz | 384.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43672-v30.xml emd-43672-v30.xml emd-43672.xml emd-43672.xml | 21.7 KB 21.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_43672.png emd_43672.png | 63.5 KB | ||
| Filedesc metadata |  emd-43672.cif.gz emd-43672.cif.gz | 7.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43672 http://ftp.pdbj.org/pub/emdb/structures/EMD-43672 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43672 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43672 | HTTPS FTP |
-Related structure data
| Related structure data |  8vynMC  8vymC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_43672.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43672.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite Map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8332 Å | ||||||||||||||||||||||||||||||||||||
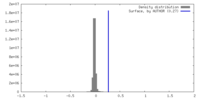
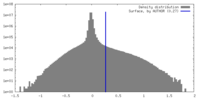
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Soluble ectodomain of human cytomegalovirus (HCMV) glycoprotein B...
| Entire | Name: Soluble ectodomain of human cytomegalovirus (HCMV) glycoprotein B (gB) stabilized in a prefusion-like conformation in complex with 1G2 and 7H3 |
|---|---|
| Components |
|
-Supramolecule #1: Soluble ectodomain of human cytomegalovirus (HCMV) glycoprotein B...
| Supramolecule | Name: Soluble ectodomain of human cytomegalovirus (HCMV) glycoprotein B (gB) stabilized in a prefusion-like conformation in complex with 1G2 and 7H3 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|
-Macromolecule #1: Envelope glycoprotein B
| Macromolecule | Name: Envelope glycoprotein B / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human betaherpesvirus 5 Human betaherpesvirus 5 |
| Molecular weight | Theoretical: 88.918586 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MESRIWCLVV CVNLCIVCLG AAVSSSSTRG TSATHSHHSS HTTSAAHSRS GSVSQRVTSS QTVSHGVNET IYNTTLKYGD VVGVNTTKY PYRVCSMAQG LDLIRFERNI VCTSMKPINE DLDEGIMVVY KRNICAHTFK VRVYQKVLTF RRSYAYIHTT Y LLGSNTEY ...String: MESRIWCLVV CVNLCIVCLG AAVSSSSTRG TSATHSHHSS HTTSAAHSRS GSVSQRVTSS QTVSHGVNET IYNTTLKYGD VVGVNTTKY PYRVCSMAQG LDLIRFERNI VCTSMKPINE DLDEGIMVVY KRNICAHTFK VRVYQKVLTF RRSYAYIHTT Y LLGSNTEY VAPPMWEIHH INSHSQCYSS YSRVIAGTVF VAYHRDSYEN KTMQLMPDDY SNTCSTRYVT VKDQWHSRGS TW LYRETSN LNCMVTITTA RSKYPYHFFI TSTGDVVDIS PFYNGTNRNA SYFGENADKF FIFPNYTIVS DFGRPNSALE THR LVAFLE RADSVISWDI QDEKNVTCQL TFWEASERTI RSEAEDSYHF SSAKMTATFL SKKQEVNMSD SALDCVRDEA INKL QQIFN TSYNQTYEKY GNVSVFETTG GLVVFWQGIK QKSLVELERL ANRSSLNLTH NSTKSSTDGN NATHLSNMES VHNLV YAQL QFTYDTLRGY INRALAQIAE AWCVDQRRTL EVFKELSKIN PSAILSAIYN KPIAARFMGD VLGLASCVTI NQTSVK VLR DMNVKESPGR CYSRPVVIFN FANSSYVQYG QLGEDNEILL GNHRTEECQL PSLKIFIAGN SAYEYVDYLF KRMIDLS SI STVDSMIALD CDPLCNTDFR VLELYSQKEL RSSNVFDLEE IMREFNSYKQ RVKYVEDKVV DPGSGYIPEA PRDGQAYV R KDGEWVLLST FLGAAASLEV LFQGPGHHHH HHHHSAWSHP QFEKGGASGG GGSGGSAWSH PQFEK UniProtKB: Envelope glycoprotein B |
-Macromolecule #2: 7H3 Fab Heavy Chain
| Macromolecule | Name: 7H3 Fab Heavy Chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.152064 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVQSGAE VKNPGASVKV SCKASGYTFT DYYIHWVRQA PGQGLEWMGW FNPNSGGTNF VQNFQGRVTM TRDTSISTAY MELSRLRSD DTAMYYCAKD SAKTASAYYG LNFFYYGMDV WGQGTTVTVS SASTKGPSVF PLAPSSKSTS GGTAALGCLV K DYFPEPVT ...String: QVQLVQSGAE VKNPGASVKV SCKASGYTFT DYYIHWVRQA PGQGLEWMGW FNPNSGGTNF VQNFQGRVTM TRDTSISTAY MELSRLRSD DTAMYYCAKD SAKTASAYYG LNFFYYGMDV WGQGTTVTVS SASTKGPSVF PLAPSSKSTS GGTAALGCLV K DYFPEPVT VSWNSGALTS GVHTFPAVLQ SSGLYSLSSV VTVPSSSLGT QTYICNVNHK PSNTKVDKKV EPKSCD |
-Macromolecule #3: 7H3 Fab Light Chain
| Macromolecule | Name: 7H3 Fab Light Chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.879361 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSVLSQPPSA SGTPGQRVTI SCSGSSSNIG KNYVYWYQQV PGTAPKLLMF KNNQRPSGVP DRFSGSKSGT SASLAISGLR SEDEADYYC SAWDGSLSRP LFGGGTKVTV LGQPKAAPSV TLFPPSSEEL QANKATLVCL ISDFYPGAVT VAWKADSSPV K AGVETTTP ...String: QSVLSQPPSA SGTPGQRVTI SCSGSSSNIG KNYVYWYQQV PGTAPKLLMF KNNQRPSGVP DRFSGSKSGT SASLAISGLR SEDEADYYC SAWDGSLSRP LFGGGTKVTV LGQPKAAPSV TLFPPSSEEL QANKATLVCL ISDFYPGAVT VAWKADSSPV K AGVETTTP SKQSNNKYAA SSYLSLTPEQ WKSHRSYSCQ VTHEGSTVEK TVAPTECS |
-Macromolecule #4: 1G2 Fab Heavy Chain
| Macromolecule | Name: 1G2 Fab Heavy Chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.206172 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QLQLQESGPG LVKPSETLSL TCTVSGASID RSTYYWGWIR QPPGKGLEWI ANIYYNGRAV YSPSLKSRVT ISVDTSKNQF SLKVRSLTA ADTAVYYCAT RWNYFFDFDY WGRGTLVTVS SASTKGPSVF PLAPSSKSTS GGTAALGCLV KDYFPEPVTV S WNSGALTS ...String: QLQLQESGPG LVKPSETLSL TCTVSGASID RSTYYWGWIR QPPGKGLEWI ANIYYNGRAV YSPSLKSRVT ISVDTSKNQF SLKVRSLTA ADTAVYYCAT RWNYFFDFDY WGRGTLVTVS SASTKGPSVF PLAPSSKSTS GGTAALGCLV KDYFPEPVTV S WNSGALTS GVHTFPAVLQ SSGLYSLSSV VTVPSSSLGT QTYICNVNHK PSNTKVDKKV EPKSCD |
-Macromolecule #5: 1G2 Fab Light Chain
| Macromolecule | Name: 1G2 Fab Light Chain / type: protein_or_peptide / ID: 5 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.853125 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSVLTQPPSA SGTPGQRVTI SCSGSSSNIE TNYVSWYQQF PGTAPKLLIY RNNQRPSGVP DRFSGSKSGT SASLAISGLR SEDEAEYYC GTWDDNSWVF GGGTKLTVLG QPKAAPSVTL FPPSSEELQA NKATLVCLIS DFYPGAVTVA WKADSSPVKA G VETTTPSK ...String: QSVLTQPPSA SGTPGQRVTI SCSGSSSNIE TNYVSWYQQF PGTAPKLLIY RNNQRPSGVP DRFSGSKSGT SASLAISGLR SEDEAEYYC GTWDDNSWVF GGGTKLTVLG QPKAAPSVTL FPPSSEELQA NKATLVCLIS DFYPGAVTVA WKADSSPVKA G VETTTPSK QSNNKYAASS YLSLTPEQWK SHRSYSCQVT HEGSTVEKTV APTECS |
-Macromolecule #8: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 8 / Number of copies: 12 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: 2 mM Tris pH 8, 200 mM NaCl, 0.02% w/v sodium azide, 3% (v/v) glycerol, 0.12% (w/v) CHAPS, 0.01% (w/v) amphipol A8-35 | |||||||||||||||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 12524 / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)