[English] 日本語
 Yorodumi
Yorodumi- EMDB-43279: Cryo-EM structure of short form insulin receptor (IR-A) with four... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of short form insulin receptor (IR-A) with four IGF2 bound, symmetric conformation. | |||||||||
 Map data Map data | Cryo-EM structure of short form insulin receptor (IR-A) with four IGF2 bound, symmetric conformation. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Insulin receptor / IGF2 / RTK / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of muscle cell differentiation / positive regulation of skeletal muscle tissue growth / embryonic placenta morphogenesis / regulation of muscle cell differentiation / Signaling by Type 1 Insulin-like Growth Factor 1 Receptor (IGF1R) / IRS-related events triggered by IGF1R / genomic imprinting / regulation of female gonad development / positive regulation of organ growth / positive regulation of meiotic cell cycle ...negative regulation of muscle cell differentiation / positive regulation of skeletal muscle tissue growth / embryonic placenta morphogenesis / regulation of muscle cell differentiation / Signaling by Type 1 Insulin-like Growth Factor 1 Receptor (IGF1R) / IRS-related events triggered by IGF1R / genomic imprinting / regulation of female gonad development / positive regulation of organ growth / positive regulation of meiotic cell cycle / transmembrane receptor protein tyrosine kinase activator activity / insulin-like growth factor II binding / positive regulation of developmental growth / male sex determination / insulin receptor complex / exocrine pancreas development / insulin-like growth factor I binding / positive regulation of multicellular organism growth / positive regulation of protein-containing complex disassembly / dendritic spine maintenance / cargo receptor activity / positive regulation of vascular endothelial cell proliferation / insulin binding / PTB domain binding / adrenal gland development / neuronal cell body membrane / Signaling by Insulin receptor / IRS activation / positive regulation of activated T cell proliferation / positive regulation of respiratory burst / amyloid-beta clearance / regulation of embryonic development / positive regulation of cell division / positive regulation of receptor internalization / protein kinase activator activity / insulin receptor substrate binding / epidermis development / embryonic placenta development / positive regulation of glycogen biosynthetic process / Signal attenuation / phosphatidylinositol 3-kinase binding / SHC-related events triggered by IGF1R / transport across blood-brain barrier / positive regulation of insulin receptor signaling pathway / heart morphogenesis / activation of protein kinase B activity / dendrite membrane / Insulin receptor recycling / insulin-like growth factor receptor binding / striated muscle cell differentiation / neuron projection maintenance / receptor-mediated endocytosis / positive regulation of mitotic nuclear division / Insulin receptor signalling cascade / positive regulation of MAP kinase activity / insulin-like growth factor receptor signaling pathway / protein serine/threonine kinase activator activity / platelet alpha granule lumen / positive regulation of glycolytic process / learning / animal organ morphogenesis / positive regulation of D-glucose import / insulin receptor binding / growth factor activity / placental growth factor receptor activity / insulin receptor activity / vascular endothelial growth factor receptor activity / hepatocyte growth factor receptor activity / macrophage colony-stimulating factor receptor activity / platelet-derived growth factor alpha-receptor activity / platelet-derived growth factor beta-receptor activity / stem cell factor receptor activity / boss receptor activity / protein tyrosine kinase collagen receptor activity / brain-derived neurotrophic factor receptor activity / transmembrane-ephrin receptor activity / GPI-linked ephrin receptor activity / epidermal growth factor receptor activity / fibroblast growth factor receptor activity / insulin-like growth factor receptor activity / receptor protein-tyrosine kinase / peptidyl-tyrosine phosphorylation / hormone activity / caveola / receptor internalization / memory / cell surface receptor protein tyrosine kinase signaling pathway / cellular response to insulin stimulus / glucose metabolic process / Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) / male gonad development / osteoblast differentiation / positive regulation of nitric oxide biosynthetic process / integrin binding / late endosome / insulin receptor signaling pathway / glucose homeostasis / Platelet degranulation / amyloid-beta binding / PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | An W / Hall C / Li J / Huang A / Wu J / Park J / Bai XC / Choi E | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Activation of the insulin receptor by insulin-like growth factor 2. Authors: Weidong An / Catherine Hall / Jie Li / Albert Hung / Jiayi Wu / Junhee Park / Liwei Wang / Xiao-Chen Bai / Eunhee Choi /  Abstract: Insulin receptor (IR) controls growth and metabolism. Insulin-like growth factor 2 (IGF2) has different binding properties on two IR isoforms, mimicking insulin's function. However, the molecular ...Insulin receptor (IR) controls growth and metabolism. Insulin-like growth factor 2 (IGF2) has different binding properties on two IR isoforms, mimicking insulin's function. However, the molecular mechanism underlying IGF2-induced IR activation remains unclear. Here, we present cryo-EM structures of full-length human long isoform IR (IR-B) in both the inactive and IGF2-bound active states, and short isoform IR (IR-A) in the IGF2-bound active state. Under saturated IGF2 concentrations, both the IR-A and IR-B adopt predominantly asymmetric conformations with two or three IGF2s bound at site-1 and site-2, which differs from that insulin saturated IR forms an exclusively T-shaped symmetric conformation. IGF2 exhibits a relatively weak binding to IR site-2 compared to insulin, making it less potent in promoting full IR activation. Cell-based experiments validated the functional importance of IGF2 binding to two distinct binding sites in optimal IR signaling and trafficking. In the inactive state, the C-terminus of α-CT of IR-B contacts FnIII-2 domain of the same protomer, hindering its threading into the C-loop of IGF2, thus reducing the association rate of IGF2 with IR-B. Collectively, our studies demonstrate the activation mechanism of IR by IGF2 and reveal the molecular basis underlying the different affinity of IGF2 to IR-A and IR-B. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43279.map.gz emd_43279.map.gz | 45 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43279-v30.xml emd-43279-v30.xml emd-43279.xml emd-43279.xml | 19.1 KB 19.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_43279.png emd_43279.png | 36.8 KB | ||
| Filedesc metadata |  emd-43279.cif.gz emd-43279.cif.gz | 6.6 KB | ||
| Others |  emd_43279_half_map_1.map.gz emd_43279_half_map_1.map.gz emd_43279_half_map_2.map.gz emd_43279_half_map_2.map.gz | 57.5 MB 57.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43279 http://ftp.pdbj.org/pub/emdb/structures/EMD-43279 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43279 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43279 | HTTPS FTP |
-Validation report
| Summary document |  emd_43279_validation.pdf.gz emd_43279_validation.pdf.gz | 758.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_43279_full_validation.pdf.gz emd_43279_full_validation.pdf.gz | 758.5 KB | Display | |
| Data in XML |  emd_43279_validation.xml.gz emd_43279_validation.xml.gz | 12.3 KB | Display | |
| Data in CIF |  emd_43279_validation.cif.gz emd_43279_validation.cif.gz | 14.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43279 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43279 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43279 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43279 | HTTPS FTP |
-Related structure data
| Related structure data |  8vjbMC  8u4bC  8u4cC  8u4eC  8vjcC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_43279.map.gz / Format: CCP4 / Size: 75.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43279.map.gz / Format: CCP4 / Size: 75.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of short form insulin receptor (IR-A) with four IGF2 bound, symmetric conformation. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
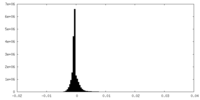
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Cryo-EM structure of short form insulin receptor (IR-A)...
| File | emd_43279_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of short form insulin receptor (IR-A) with four IGF2 bound, symmetric conformation. Half map 1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
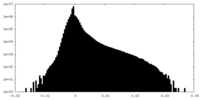
| Density Histograms |
-Half map: Cryo-EM structure of short form insulin receptor (IR-A)...
| File | emd_43279_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of short form insulin receptor (IR-A) with four IGF2 bound, symmetric conformation. Half map 2. | ||||||||||||
| Projections & Slices |
| ||||||||||||
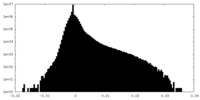
| Density Histograms |
- Sample components
Sample components
-Entire : Short form insulin receptor (IR-A) with four IGF2 bound, symmetri...
| Entire | Name: Short form insulin receptor (IR-A) with four IGF2 bound, symmetric conformation. |
|---|---|
| Components |
|
-Supramolecule #1: Short form insulin receptor (IR-A) with four IGF2 bound, symmetri...
| Supramolecule | Name: Short form insulin receptor (IR-A) with four IGF2 bound, symmetric conformation. type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Isoform Short of Insulin receptor
| Macromolecule | Name: Isoform Short of Insulin receptor / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: receptor protein-tyrosine kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 155.329094 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MATGGRRGAA AAPLLVAVAA LLLGAAGHLY PGEVCPGMDI RNNLTRLHEL ENCSVIEGHL QILLMFKTRP EDFRDLSFPK LIMITDYLL LFRVYGLESL KDLFPNLTVI RGSRLFFNYA LVIFEMVHLK ELGLYNLMNI TRGSVRIEKN NELCYLATID W SRILDSVE ...String: MATGGRRGAA AAPLLVAVAA LLLGAAGHLY PGEVCPGMDI RNNLTRLHEL ENCSVIEGHL QILLMFKTRP EDFRDLSFPK LIMITDYLL LFRVYGLESL KDLFPNLTVI RGSRLFFNYA LVIFEMVHLK ELGLYNLMNI TRGSVRIEKN NELCYLATID W SRILDSVE DNYIVLNKDD NEECGDICPG TAKGKTNCPA TVINGQFVER CWTHSHCQKV CPTICKSHGC TAEGLCCHSE CL GNCSQPD DPTKCVACRN FYLDGRCVET CPPPYYHFQD WRCVNFSFCQ DLHHKCKNSR RQGCHQYVIH NNKCIPECPS GYT MNSSNL LCTPCLGPCP KVCHLLEGEK TIDSVTSAQE LRGCTVINGS LIINIRGGNN LAAELEANLG LIEEISGYLK IRRS YALVS LSFFRKLRLI RGETLEIGNY SFYALDNQNL RQLWDWSKHN LTITQGKLFF HYNPKLCLSE IHKMEEVSGT KGRQE RNDI ALKTNGDQAS CENELLKFSY IRTSFDKILL RWEPYWPPDF RDLLGFMLFY KEAPYQNVTE FDGQDACGSN SWTVVD IDP PLRSNDPKSQ NHPGWLMRGL KPWTQYAIFV KTLVTFSDER RTYGAKSDII YVQTDATNPS VPLDPISVSN SSSQIIL KW KPPSDPNGNI THYLVFWERQ AEDSELFELD YCLKGLKLPS RTWSPPFESE DSQKHNQSEY EDSAGECCSC PKTDSQIL K ELEESSFRKT FEDYLHNVVF VPRPSRKRRS LGDVGNVTVA VPTVAAFPNT SSTSVPTSPE EHRPFEKVVN KESLVISGL RHFTGYRIEL QACNQDTPEE RCSVAAYVSA RTMPEAKADD IVGPVTHEIF ENNVVHLMWQ EPKEPNGLIV LYEVSYRRYG DEELHLCVS RKHFALERGC RLRGLSPGNY SVRIRATSLA GNGSWTEPTY FYVTDYLDVP SNIAKIIIGP LIFVFLFSVV I GSIYLFLR KRQPDGPLGP LYASSNPEYL SASDVFPCSV YVPDEWEVSR EKITLLRELG QGSFGMVYEG NARDIIKGEA ET RVAVKTV NESASLRERI EFLNEASVMK GFTCHHVVRL LGVVSKGQPT LVVMELMAHG DLKSYLRSLR PEAENNPGRP PPT LQEMIQ MAAEIADGMA YLNAKKFVHR DLAARNCMVA HDFTVKIGDF GMTRDIYETD YYRKGGKGLL PVRWMAPESL KDGV FTTSS DMWSFGVVLW EITSLAEQPY QGLSNEQVLK FVMDGGYLDQ PDNCPERVTD LMRMCWQFNP KMRPTFLEIV NLLKD DLHP SFPEVSFFHS EENKAPESEE LEMEFEDMEN VPLDRSSHCQ REEAGGRDGG SSLGFKRSYE EHIPYTHMNG GKKNGR ILT LPRSNPS UniProtKB: Insulin receptor |
-Macromolecule #2: Insulin-like growth factor II
| Macromolecule | Name: Insulin-like growth factor II / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 20.170398 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGIPMGKSML VLLTFLAFAS CCIAAYRPSE TLCGGELVDT LQFVCGDRGF YFSRPASRVS RRSRGIVEEC CFRSCDLALL ETYCATPAK SERDVSTPPT VLPDNFPRYP VGKFFQYDTW KQSTQRLRRG LPALLRARRG HVLAKELEAF REAKRHRPLI A LPTQDPAH GGAPPEMASN RK UniProtKB: Insulin-like growth factor 2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.6 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-8vjb: |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)