[English] 日本語
 Yorodumi
Yorodumi- EMDB-41149: Cryo-EM Structure of Spike Glycoprotein from Bat Coronavirus WIV1... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of Spike Glycoprotein from Bat Coronavirus WIV1 in Closed Conformation | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Spike / Glycoprotein / Coronavirus / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationendocytosis involved in viral entry into host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / receptor-mediated virion attachment to host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / host cell plasma membrane / virion membrane / membrane Similarity search - Function | |||||||||
| Biological species |  Bat SARS-like coronavirus WIV1 Bat SARS-like coronavirus WIV1 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 1.88 Å | |||||||||
 Authors Authors | Bostina M / Hills FR / Eruera A | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2024 Journal: PLoS Pathog / Year: 2024Title: Variation in structural motifs within SARS-related coronavirus spike proteins. Authors: Francesca R Hills / Alice-Roza Eruera / James Hodgkinson-Bean / Fátima Jorge / Richard Easingwood / Simon H J Brown / James C Bouwer / Yi-Ping Li / Laura N Burga / Mihnea Bostina /    Abstract: SARS-CoV-2 is the third known coronavirus (CoV) that has crossed the animal-human barrier in the last two decades. However, little structural information exists related to the close genetic species ...SARS-CoV-2 is the third known coronavirus (CoV) that has crossed the animal-human barrier in the last two decades. However, little structural information exists related to the close genetic species within the SARS-related coronaviruses. Here, we present three novel SARS-related CoV spike protein structures solved by single particle cryo-electron microscopy analysis derived from bat (bat SL-CoV WIV1) and civet (cCoV-SZ3, cCoV-007) hosts. We report complex glycan trees that decorate the glycoproteins and density for water molecules which facilitated modeling of the water molecule coordination networks within structurally important regions. We note structural conservation of the fatty acid binding pocket and presence of a linoleic acid molecule which are associated with stabilization of the receptor binding domains in the "down" conformation. Additionally, the N-terminal biliverdin binding pocket is occupied by a density in all the structures. Finally, we analyzed structural differences in a loop of the receptor binding motif between coronaviruses known to infect humans and the animal coronaviruses described in this study, which regulate binding to the human angiotensin converting enzyme 2 receptor. This study offers a structural framework to evaluate the close relatives of SARS-CoV-2, the ability to inform pandemic prevention, and aid in the development of pan-neutralizing treatments. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41149.map.gz emd_41149.map.gz | 175.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41149-v30.xml emd-41149-v30.xml emd-41149.xml emd-41149.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41149.png emd_41149.png | 59.5 KB | ||
| Filedesc metadata |  emd-41149.cif.gz emd-41149.cif.gz | 6.4 KB | ||
| Others |  emd_41149_half_map_1.map.gz emd_41149_half_map_1.map.gz emd_41149_half_map_2.map.gz emd_41149_half_map_2.map.gz | 322.5 MB 322.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41149 http://ftp.pdbj.org/pub/emdb/structures/EMD-41149 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41149 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41149 | HTTPS FTP |
-Validation report
| Summary document |  emd_41149_validation.pdf.gz emd_41149_validation.pdf.gz | 761.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_41149_full_validation.pdf.gz emd_41149_full_validation.pdf.gz | 761 KB | Display | |
| Data in XML |  emd_41149_validation.xml.gz emd_41149_validation.xml.gz | 17.1 KB | Display | |
| Data in CIF |  emd_41149_validation.cif.gz emd_41149_validation.cif.gz | 20.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41149 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41149 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41149 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41149 | HTTPS FTP |
-Related structure data
| Related structure data |  8tc0MC  8tc1C  8tc5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41149.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41149.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_41149_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
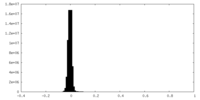
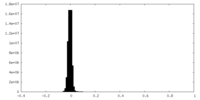
| Density Histograms |
-Half map: #1
| File | emd_41149_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
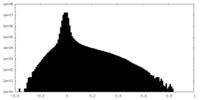
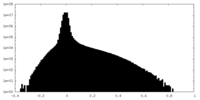
| Density Histograms |
- Sample components
Sample components
-Entire : Bat SARS-like coronavirus WIV1
| Entire | Name:  Bat SARS-like coronavirus WIV1 Bat SARS-like coronavirus WIV1 |
|---|---|
| Components |
|
-Supramolecule #1: Bat SARS-like coronavirus WIV1
| Supramolecule | Name: Bat SARS-like coronavirus WIV1 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1 / NCBI-ID: 1415852 / Sci species name: Bat SARS-like coronavirus WIV1 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bat SARS-like coronavirus WIV1 Bat SARS-like coronavirus WIV1 |
| Molecular weight | Theoretical: 122.841266 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: KCLDFDDRTP PANTQFLSSH RGVYYPDDIF RSNVLHLVQD HFLPFDSNVT RFITFGLNFD NPIIPFKDGI YFAATEKSNV IRGWVFGST MNNKSQSVII MNNSTNLVIR ACNFELCDNP FFVVLKSNNT QIPSYIFNNA FNCTFEYVSK DFNLDLGEKP G NFKDLREF ...String: KCLDFDDRTP PANTQFLSSH RGVYYPDDIF RSNVLHLVQD HFLPFDSNVT RFITFGLNFD NPIIPFKDGI YFAATEKSNV IRGWVFGST MNNKSQSVII MNNSTNLVIR ACNFELCDNP FFVVLKSNNT QIPSYIFNNA FNCTFEYVSK DFNLDLGEKP G NFKDLREF VFRNKDGFLH VYSGYQPISA ASGLPTGFNA LKPIFKLPLG INITNFRTLL TAFPPRPDYW GTSAAAYFVG YL KPTTFML KYDENGTITD AVDCSQNPLA ELKCSVKSFE IDKGIYQTSN FRVAPSKEVV RFPNITNLCP FGEVFNATTF PSV YAWERK RISNCVADYS VLYNSTSFST FKCYGVSATK LNDLCFSNVY ADSFVVKGDD VRQIAPGQTG VIADYNYKLP DDFT GCVLA WNTRNIDATQ TGNYNYKYRS LRHGKLRPFE RDISNVPFSP DGKPCTPPAF NCYWPLNDYG FYITNGIGYQ PYRVV VLSF ELLNAPATVC GPKLSTDLIK NQCVNFNFNG LTGTGVLTPS SKRFQPFQQF GRDVSDFTDS VRDPKTSEIL DISPCS FGG VSVITPGTNT SSEVAVLYQD VNCTDVPVAI HADQLTPSWR VHSTGNNVFQ TQAGCLIGAE HVDTSYECDI PIGAGIC AS YHTVSSLRST SQKSIVAYTM SLGADSSIAY SNNTIAIPTN FSISITTEVM PVSMAKTSVD CNMYICGDST ECANLLLQ Y GSFCTQLNRA LSGIAVEQDR NTREVFAQVK QMYKTPTLKD FGGFNFSQIL PDPLKPTKRS FIEDLLFNKV TLADAGFMK QYGECLGDIN ARDLICAQKF NGLTVLPPLL TDDMIAAYTA ALVSGTATAG WTFGAGAALQ IPFAMQMAYR FNGIGVTQNV LYENQKQIA NQFNKAISQI QESLTTTSTA LGKLQDVVNQ NAQALNTLVK QLSSNFGAIS SVLNDILSRL DKVEAEVQID R LITGRLQS LQTYVTQQLI RAAEIRASAN LAATKMSECV LGQSKRVDFC GKGYHLMSFP QAAPHGVVFL HVTYVPSQER NF TTAPAIC HEGKAYFPRE GVFVFNGTSW FITQRNFFSP QIITTDNTFV SGSCDVVIGI INNTVYDPLQ PE UniProtKB: Spike glycoprotein |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 15 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #7: LINOLEIC ACID
| Macromolecule | Name: LINOLEIC ACID / type: ligand / ID: 7 / Number of copies: 3 / Formula: EIC |
|---|---|
| Molecular weight | Theoretical: 280.445 Da |
| Chemical component information |  ChemComp-EIC: |
-Macromolecule #8: water
| Macromolecule | Name: water / type: ligand / ID: 8 / Number of copies: 207 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 67.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: OTHER / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 1.88 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 800000 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)