+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 DMV nsp3-4 pore complex (extended-pore) | |||||||||
 Map data Map data | local resolution map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Double membrane vesicle / pore complex / nsp3 / nsp4 / RNA transport / VIRAL PROTEIN | |||||||||
| Biological species |  | |||||||||
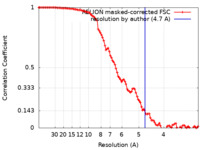
| Method | subtomogram averaging / cryo EM / Resolution: 4.7 Å | |||||||||
 Authors Authors | Huang YX / Zhong LJ / Zhang WX / Ni T | |||||||||
| Funding support |  Hong Kong, 1 items Hong Kong, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Molecular architecture of coronavirus double-membrane vesicle pore complex. Authors: Yixin Huang / Tongyun Wang / Lijie Zhong / Wenxin Zhang / Yu Zhang / Xiulian Yu / Shuofeng Yuan / Tao Ni /  Abstract: Coronaviruses remodel the intracellular host membranes during replication, forming double-membrane vesicles (DMVs) to accommodate viral RNA synthesis and modifications. SARS-CoV-2 non-structural ...Coronaviruses remodel the intracellular host membranes during replication, forming double-membrane vesicles (DMVs) to accommodate viral RNA synthesis and modifications. SARS-CoV-2 non-structural protein 3 (nsp3) and nsp4 are the minimal viral components required to induce DMV formation and to form a double-membrane-spanning pore, essential for the transport of newly synthesized viral RNAs. The mechanism of DMV pore complex formation remains unknown. Here we describe the molecular architecture of the SARS-CoV-2 nsp3-nsp4 pore complex, as resolved by cryogenic electron tomography and subtomogram averaging in isolated DMVs. The structures uncover an unexpected stoichiometry and topology of the nsp3-nsp4 pore complex comprising 12 copies each of nsp3 and nsp4, organized in 4 concentric stacking hexamer rings, mimicking a miniature nuclear pore complex. The transmembrane domains are interdigitated to create a high local curvature at the double-membrane junction, coupling double-membrane reorganization with pore formation. The ectodomains form extensive contacts in a pseudo-12-fold symmetry, belting the pore complex from the intermembrane space. A central positively charged ring of arginine residues coordinates the putative RNA translocation, essential for virus replication. Our work establishes a framework for understanding DMV pore formation and RNA translocation, providing a structural basis for the development of new antiviral strategies to combat coronavirus infection. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_39111.map.gz emd_39111.map.gz | 51.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-39111-v30.xml emd-39111-v30.xml emd-39111.xml emd-39111.xml | 15.4 KB 15.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_39111_fsc.xml emd_39111_fsc.xml | 10.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_39111.png emd_39111.png | 104.4 KB | ||
| Masks |  emd_39111_msk_1.map emd_39111_msk_1.map | 91.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-39111.cif.gz emd-39111.cif.gz | 4.6 KB | ||
| Others |  emd_39111_half_map_1.map.gz emd_39111_half_map_1.map.gz emd_39111_half_map_2.map.gz emd_39111_half_map_2.map.gz | 43.6 MB 43.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39111 http://ftp.pdbj.org/pub/emdb/structures/EMD-39111 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39111 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39111 | HTTPS FTP |
-Validation report
| Summary document |  emd_39111_validation.pdf.gz emd_39111_validation.pdf.gz | 908.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_39111_full_validation.pdf.gz emd_39111_full_validation.pdf.gz | 908.1 KB | Display | |
| Data in XML |  emd_39111_validation.xml.gz emd_39111_validation.xml.gz | 17.6 KB | Display | |
| Data in CIF |  emd_39111_validation.cif.gz emd_39111_validation.cif.gz | 23.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39111 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39111 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39111 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39111 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_39111.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_39111.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | local resolution map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size |
| ||||||||||||||||||||||||||||||||||||
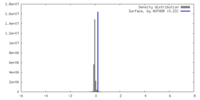
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_39111_msk_1.map emd_39111_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
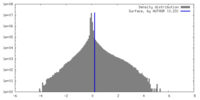
| Density Histograms |
-Half map: #2
| File | emd_39111_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
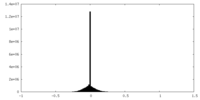
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 nsp3-4 pore complex
| Entire | Name: SARS-CoV-2 nsp3-4 pore complex |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 nsp3-4 pore complex
| Supramolecule | Name: SARS-CoV-2 nsp3-4 pore complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
Details: 150mM NaCl, 10mM Tris-HCl, 1mM EDTA | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: EMS Lacey Carbon / Support film - Material: CARBON | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: TFS FALCON 4i (4k x 4k) / Average electron dose: 3.0 e/Å2 Details: The tilt-series were acquired using Thermofisher Krios equipped with a Falcon 4i camera and Selectris energy filter. A dose-symmetric scheme (group of 2) was used, with a tilt range of -51 ...Details: The tilt-series were acquired using Thermofisher Krios equipped with a Falcon 4i camera and Selectris energy filter. A dose-symmetric scheme (group of 2) was used, with a tilt range of -51 to 51 (or -60 to 60) at 3 increments and an exposure dose of 3 e-/A2 per image. The total dose was 105 (or 123) e-/A2. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 6.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)