[English] 日本語
 Yorodumi
Yorodumi- EMDB-38978: The focused refinement map (G-protein) of WN561 bound APLNR-Gi1 s... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The focused refinement map (G-protein) of WN561 bound APLNR-Gi1 structure | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | APLNR / WN561 / STRUCTURAL PROTEIN | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
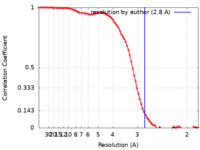
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Wang W / Ji S / Zhang Y | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2024 Journal: Cell / Year: 2024Title: Structure-based design of non-hypertrophic apelin receptor modulator. Authors: Wei-Wei Wang / Su-Yu Ji / Wenjia Zhang / Junxia Zhang / Chenxi Cai / Rubi Hu / Shao-Kun Zang / Luwei Miao / Haomang Xu / Li-Nan Chen / Zongkuai Yang / Jia Guo / Jiao Qin / Dan-Dan Shen / ...Authors: Wei-Wei Wang / Su-Yu Ji / Wenjia Zhang / Junxia Zhang / Chenxi Cai / Rubi Hu / Shao-Kun Zang / Luwei Miao / Haomang Xu / Li-Nan Chen / Zongkuai Yang / Jia Guo / Jiao Qin / Dan-Dan Shen / Ping Liang / Yan Zhang / Yan Zhang /  Abstract: Apelin is a key hormone in cardiovascular homeostasis that activates the apelin receptor (APLNR), which is regarded as a promising therapeutic target for cardiovascular disease. However, adverse ...Apelin is a key hormone in cardiovascular homeostasis that activates the apelin receptor (APLNR), which is regarded as a promising therapeutic target for cardiovascular disease. However, adverse effects through the β-arrestin pathway limit its pharmacological use. Here, we report cryoelectron microscopy (cryo-EM) structures of APLNR-G complexes bound to three agonists with divergent signaling profiles. Combined with functional assays, we have identified "twin hotspots" in APLNR as key determinants for signaling bias, guiding the rational design of two exclusive G-protein-biased agonists WN353 and WN561. Cryo-EM structures of WN353- and WN561-stimulated APLNR-G protein complexes further confirm that the designed ligands adopt the desired poses. Pathophysiological experiments have provided evidence that WN561 demonstrates superior therapeutic effects against cardiac hypertrophy and reduced adverse effects compared with the established APLNR agonists. In summary, our designed APLNR modulator may facilitate the development of next-generation cardiovascular medications. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38978.map.gz emd_38978.map.gz | 33.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38978-v30.xml emd-38978-v30.xml emd-38978.xml emd-38978.xml | 13 KB 13 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_38978_fsc.xml emd_38978_fsc.xml | 10.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_38978.png emd_38978.png | 49.1 KB | ||
| Masks |  emd_38978_msk_1.map emd_38978_msk_1.map | 42.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-38978.cif.gz emd-38978.cif.gz | 4.5 KB | ||
| Others |  emd_38978_half_map_1.map.gz emd_38978_half_map_1.map.gz emd_38978_half_map_2.map.gz emd_38978_half_map_2.map.gz | 33.1 MB 33.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38978 http://ftp.pdbj.org/pub/emdb/structures/EMD-38978 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38978 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38978 | HTTPS FTP |
-Validation report
| Summary document |  emd_38978_validation.pdf.gz emd_38978_validation.pdf.gz | 926.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_38978_full_validation.pdf.gz emd_38978_full_validation.pdf.gz | 926.2 KB | Display | |
| Data in XML |  emd_38978_validation.xml.gz emd_38978_validation.xml.gz | 13.5 KB | Display | |
| Data in CIF |  emd_38978_validation.cif.gz emd_38978_validation.cif.gz | 18.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38978 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38978 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38978 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38978 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_38978.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38978.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.93 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_38978_msk_1.map emd_38978_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_38978_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_38978_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Gi protein
| Entire | Name: Gi protein |
|---|---|
| Components |
|
-Supramolecule #1: Gi protein
| Supramolecule | Name: Gi protein / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Gi protein
| Macromolecule | Name: Gi protein / type: other / ID: 1 / Classification: other |
|---|---|
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKNTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMGR LKIDFGDSAR ADDARQLFVL AGAAEEGFMT AELAGVIKRL WKDSGVQACF NRSREYQLND SAAYYLNDLD RIAQPNYIPT ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKNTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMGR LKIDFGDSAR ADDARQLFVL AGAAEEGFMT AELAGVIKRL WKDSGVQACF NRSREYQLND SAAYYLNDLD RIAQPNYIPT QQDVLRTRVK TTGIVETHFT FKDLHFKMFD VGAQRSERKK WIHCFEGVTA IIFCVALSDY DLVLAEDEEM NRMHASMKLF DSICNNKWFT DTSIILFLNK KDLFEEKIKK SPLTICYPEY AGSNTYEEAA AYIQCQFEDL NKRKDTKEIY THFTCSTDTK NVQFVFDAVT DVIIKNNLKD CGLFSELDQL RQEAEQLKNQ IRDARKACAD ATLSQITNNI DPVGRIQMRT RRTLRGHLAK IYAMHWGTDS RLLVSASQDG KLIIWDSYTT NKVHAIPLRS SWVMTCAYAP SGNYVACGGL DNICSIYNLK TREGNVRVSR ELAGHTGYLS CCRFLDDNQI VTSSGDTTCA LWDIETGQQT TTFTGHTGDV MSLSLAPDTR LFVSGACDAS AKLWDVREGM CRQTFTGHES DINAICFFPN GNAFATGSDD ATCRLFDLRA DQELMTYSHD NIICGITSVS FSKSGRLLLA GYDDFNCNVW DALKADRAGV LAGHDNRVSC LGVTDDGMAV ATGSWDSFLK IWNMASNNTA SIAQARKLVE QLKMEANIDR IKVSKAAADL MAYCEAHAKE DPLLTPVPAS ENPFREKKFF CAIL |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)