[English] 日本語
 Yorodumi
Yorodumi- EMDB-38081: Conventional Reconstruction of the MC-45 de novo processed riboso... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Conventional Reconstruction of the MC-45 de novo processed ribosome 50S | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ribosome 50S / RIBOSOME | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / Resolution: 3.16 Å | |||||||||
 Authors Authors | Zhang XZ / Zhu DJ / Cao WL | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2024 Journal: Sci Adv / Year: 2024Title: Correction of preferred orientation-induced distortion in cryo-electron microscopy maps. Authors: Dongjie Zhu / Weili Cao / Junxi Li / Chunling Wu / Duanfang Cao / Xinzheng Zhang /  Abstract: Reconstruction maps of cryo-electron microscopy (cryo-EM) exhibit distortion when the cryo-EM dataset is incomplete, usually caused by unevenly distributed orientations. Prior efforts had been ...Reconstruction maps of cryo-electron microscopy (cryo-EM) exhibit distortion when the cryo-EM dataset is incomplete, usually caused by unevenly distributed orientations. Prior efforts had been attempted to address this preferred orientation problem using tilt-collection strategy and modifications to grids or to air-water interfaces. However, these approaches often require time-consuming experiments, and the effect was always protein dependent. Here, we developed a procedure containing removing misaligned particles and an iterative reconstruction method based on signal-to-noise ratio of Fourier component to correct this distortion by recovering missing data using a purely computational algorithm. This procedure called signal-to-noise ratio iterative reconstruction method (SIRM) was applied on incomplete datasets of various proteins to fix distortion in cryo-EM maps and to a more isotropic resolution. In addition, SIRM provides a better reference map for further reconstruction refinements, resulting in an improved alignment, which ultimately improves map quality and benefits model building. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38081.map.gz emd_38081.map.gz | 204.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38081-v30.xml emd-38081-v30.xml emd-38081.xml emd-38081.xml | 12.3 KB 12.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_38081.png emd_38081.png | 75.7 KB | ||
| Masks |  emd_38081_msk_1.map emd_38081_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-38081.cif.gz emd-38081.cif.gz | 3.8 KB | ||
| Others |  emd_38081_half_map_1.map.gz emd_38081_half_map_1.map.gz emd_38081_half_map_2.map.gz emd_38081_half_map_2.map.gz | 200.7 MB 200.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38081 http://ftp.pdbj.org/pub/emdb/structures/EMD-38081 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38081 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38081 | HTTPS FTP |
-Validation report
| Summary document |  emd_38081_validation.pdf.gz emd_38081_validation.pdf.gz | 1.3 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_38081_full_validation.pdf.gz emd_38081_full_validation.pdf.gz | 1.3 MB | Display | |
| Data in XML |  emd_38081_validation.xml.gz emd_38081_validation.xml.gz | 15.8 KB | Display | |
| Data in CIF |  emd_38081_validation.cif.gz emd_38081_validation.cif.gz | 18.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38081 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38081 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38081 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38081 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_38081.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38081.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.94666 Å | ||||||||||||||||||||||||||||||||||||
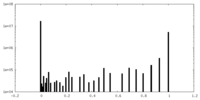
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_38081_msk_1.map emd_38081_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
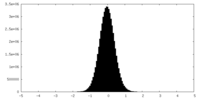
| Density Histograms |
-Half map: #2
| File | emd_38081_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
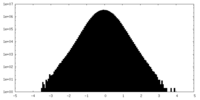
| Density Histograms |
-Half map: #1
| File | emd_38081_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
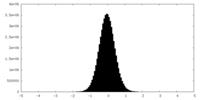
| Density Histograms |
- Sample components
Sample components
-Entire : E. coli 70S ribosome, from EMPIAR-10509
| Entire | Name: E. coli 70S ribosome, from EMPIAR-10509 |
|---|---|
| Components |
|
-Supramolecule #1: E. coli 70S ribosome, from EMPIAR-10509
| Supramolecule | Name: E. coli 70S ribosome, from EMPIAR-10509 / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
 Processing Processing | single particle reconstruction |
|---|---|
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 Details: We used EMPIAR-10509. This repository was not recorded by us. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP EMDB ID: |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.16 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 3.3) / Number images used: 38800 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. 3.3) |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)