[English] 日本語
 Yorodumi
Yorodumi- EMDB-36867: The "5+1" heteromeric structure of Lon protease consisting of a s... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The "5+1" heteromeric structure of Lon protease consisting of a spiral pentamer with Y224S mutation and an N-terminal-truncated monomeric E613K mutant | |||||||||
 Map data Map data | heteromeric 5 1 complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | AAA+ protein / ATPase / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationendopeptidase La / protein quality control for misfolded or incompletely synthesized proteins / ATP-dependent peptidase activity / cellular response to heat / sequence-specific DNA binding / serine-type endopeptidase activity / ATP hydrolysis activity / ATP binding / identical protein binding / metal ion binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Meiothermus taiwanensis (bacteria) Meiothermus taiwanensis (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.42 Å | |||||||||
 Authors Authors | Li S / Hsieh KY / Kuo CI / Zhang K / Chang CI | |||||||||
| Funding support |  Taiwan, 2 items Taiwan, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine. Authors: Shanshan Li / Kan-Yen Hsieh / Chiao-I Kuo / Tzu-Chi Lin / Szu-Hui Lee / Yi-Ru Chen / Chun-Hsiung Wang / Meng-Ru Ho / See-Yeun Ting / Kaiming Zhang / Chung-I Chang /   Abstract: Many AAA+ (ATPases associated with diverse cellular activities) proteins function as protein or DNA remodelers by threading the substrate through the central pore of their hexameric assemblies. In ...Many AAA+ (ATPases associated with diverse cellular activities) proteins function as protein or DNA remodelers by threading the substrate through the central pore of their hexameric assemblies. In this ATP-dependent translocating state, the substrate is gripped by the pore loops of the ATPase domains arranged in a universal right-handed spiral staircase organization. However, the process by which a AAA+ protein is activated to adopt this substrate-pore-loop arrangement remains unknown. We show here, using cryo-electron microscopy (cryo-EM), that the activation process of the Lon AAA+ protease may involve a pentameric assembly and a substrate-dependent incorporation of the sixth protomer to form the substrate-pore-loop contacts seen in the translocating state. Based on the structural results, we design truncated monomeric mutants that inhibit Lon activity by binding to the native pentamer and demonstrated that expressing these monomeric mutants in Escherichia coli cells containing functional Lon elicits specific phenotypes associated with lon deficiency, including the inhibition of persister cell formation. These findings uncover a substrate-dependent assembly process for the activation of a AAA+ protein and demonstrate a targeted approach to selectively inhibit its function within cells. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36867.map.gz emd_36867.map.gz | 62.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36867-v30.xml emd-36867-v30.xml emd-36867.xml emd-36867.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36867_fsc.xml emd_36867_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_36867.png emd_36867.png | 55.2 KB | ||
| Filedesc metadata |  emd-36867.cif.gz emd-36867.cif.gz | 6.2 KB | ||
| Others |  emd_36867_half_map_1.map.gz emd_36867_half_map_1.map.gz emd_36867_half_map_2.map.gz emd_36867_half_map_2.map.gz | 115.9 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36867 http://ftp.pdbj.org/pub/emdb/structures/EMD-36867 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36867 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36867 | HTTPS FTP |
-Validation report
| Summary document |  emd_36867_validation.pdf.gz emd_36867_validation.pdf.gz | 1004.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36867_full_validation.pdf.gz emd_36867_full_validation.pdf.gz | 1004.3 KB | Display | |
| Data in XML |  emd_36867_validation.xml.gz emd_36867_validation.xml.gz | 19.1 KB | Display | |
| Data in CIF |  emd_36867_validation.cif.gz emd_36867_validation.cif.gz | 24.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36867 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36867 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36867 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36867 | HTTPS FTP |
-Related structure data
| Related structure data |  8k3yMC  7yphC  7ypiC  7ypjC  7ypkC  7yuhC  7yumC  7yupC  7yutC  7yuuC  7yuvC  7yuwC  7yuxC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36867.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36867.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | heteromeric 5 1 complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.061 Å | ||||||||||||||||||||||||||||||||||||
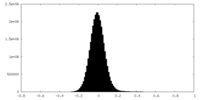
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map B
| File | emd_36867_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
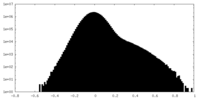
| Density Histograms |
-Half map: half map A
| File | emd_36867_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
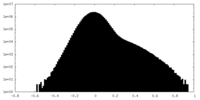
| Density Histograms |
- Sample components
Sample components
-Entire : Spiral pentamer of Lon protease with a Y224S mutation in complex ...
| Entire | Name: Spiral pentamer of Lon protease with a Y224S mutation in complex with the N-terminal-truncated monomeric E613K mutant of Lon. |
|---|---|
| Components |
|
-Supramolecule #1: Spiral pentamer of Lon protease with a Y224S mutation in complex ...
| Supramolecule | Name: Spiral pentamer of Lon protease with a Y224S mutation in complex with the N-terminal-truncated monomeric E613K mutant of Lon. type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Meiothermus taiwanensis (bacteria) Meiothermus taiwanensis (bacteria) |
-Macromolecule #1: Lon protease
| Macromolecule | Name: Lon protease / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO / EC number: endopeptidase La |
|---|---|
| Source (natural) | Organism:  Meiothermus taiwanensis (bacteria) Meiothermus taiwanensis (bacteria) |
| Molecular weight | Theoretical: 89.307383 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MRLELPVIPL RNTVILPHTT TPVDVGRAKS KRAVEEAMGA DRLIFLVAQR DPEVDDPAPD DLYTWGVQAV VKQAMRLPDG TLQVMVEAR ARAQVTDYIP GPYLRARGEV FSEIFPIDEA VVRVLVEELK EAFEKYVANH KSLRLDRYQL EAVKGTSDPA M LADTIAYH ...String: MRLELPVIPL RNTVILPHTT TPVDVGRAKS KRAVEEAMGA DRLIFLVAQR DPEVDDPAPD DLYTWGVQAV VKQAMRLPDG TLQVMVEAR ARAQVTDYIP GPYLRARGEV FSEIFPIDEA VVRVLVEELK EAFEKYVANH KSLRLDRYQL EAVKGTSDPA M LADTIAYH ATWTVAEKQE ILELTDLEAR LKKVLGLLSR DLERFELDKR VAQRVKEQMD TNQRESYLRE QMKAIQKELG GE DGLSDLE ALRKKIEEVG MPEAVKTKAL KELDRLERMQ QGSPEATVAR TYLDWLTEVP WSKADPEVLD INHTRQVLDE DHY GLKDVK ERILEYLAVR QLTQGLDVRN KAPILVLVGP PGVGKTSLGR SIARSMNRKF HRISLGGVRD EAEIRGHRRT YIGA MPGKL IHAMKQVGVI NPVILLDEID KMSSDWRGDP ASAMLEVLDP EQNNTFTDHY LDVPYDLSKV FFITTANTLQ TIPRP LLDR MEVIEIPGYT NMEKQAIARQ YLWPKQVRES GMEGRIEVTD AAILRVISEY TREAGVRGLE RELGKIARKG AKFWLE GAW EGLRTIDASD IPTYLGIPRY RPDKAETEPQ VGTAQGLAWT PVGGTLLTIE VAAVPGSGKL SLTGQLGEVM KESAQAA LT YLRAHTQDYG LPEDFYNKVD LHVHVPDGAT PKDGPSAGIT MATAIASALS RRPARMDIAM TGEVSLRGKV MPIGGVKE K LLAAHQAGIH KIVLPKDNEA QLEELPKEVL EGLEIKLVED VGEVLEYLLL PEPTMPPVVQ PSDNRQQPGA GAHHHHHH UniProtKB: Lon protease |
-Macromolecule #2: Lon protease
| Macromolecule | Name: Lon protease / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: endopeptidase La |
|---|---|
| Source (natural) | Organism:  Meiothermus taiwanensis (bacteria) Meiothermus taiwanensis (bacteria) |
| Molecular weight | Theoretical: 63.185266 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HHHHHHSSGE NLYFQGHMGL SDLEALRKKI EEVGMPEAVK TKALKELDRL ERMQQGSPEA TVARTYLDWL TEVPWSKADP EVLDINHTR QVLDEDHYGL KDVKERILEY LAVRQLTQGL DVRNKAPILV LVGPPGVGKT SLGRSIARSM NRKFHRISLG G VRDEAEIR ...String: HHHHHHSSGE NLYFQGHMGL SDLEALRKKI EEVGMPEAVK TKALKELDRL ERMQQGSPEA TVARTYLDWL TEVPWSKADP EVLDINHTR QVLDEDHYGL KDVKERILEY LAVRQLTQGL DVRNKAPILV LVGPPGVGKT SLGRSIARSM NRKFHRISLG G VRDEAEIR GHRRTYIGAM PGKLIHAMKQ VGVINPVILL DEIDKMSSDW RGDPASAMLE VLDPEQNNTF TDHYLDVPYD LS KVFFITT ANTLQTIPRP LLDRMEVIEI PGYTNMEKQA IARQYLWPKQ VRESGMEGRI EVTDAAILRV ISEYTREAGV RGL ERELGK IARKGAKFWL EGAWEGLRTI DASDIPTYLG IPRYRPDKAE TEPQVGTAQG LAWTPVGGTL LTIKVAAVPG SGKL SLTGQ LGEVMKESAQ AALTYLRAHT QDYGLPEDFY NKVDLHVHVP DGATPKDGPS AGITMATAIA SALSRRPARM DIAMT GEVS LRGKVMPIGG VKEKLLAAHQ AGIHKIVLPK DNEAQLEELP KEVLEGLEIK LVEDVGEVLE YLLLPEPTMP PVVQPS DNR QQPGAGA UniProtKB: Lon protease |
-Macromolecule #3: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 4 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.7 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: 20 mM Tris-HCl, 200 mM NaCl, 10 mM MgCl2, 1 mM DTT | |||||||||||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.4000000000000001 µm / Nominal magnification: 81000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)