[English] 日本語
 Yorodumi
Yorodumi- EMDB-36688: alpha-Hemolysin(G122S/K147R)-SpyTag/SpyCatcher head to head 14-mer -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | alpha-Hemolysin(G122S/K147R)-SpyTag/SpyCatcher head to head 14-mer | |||||||||
 Map data Map data | B-factor Sharpened map with B-factor value of -60 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | pore / chimera protein / spy-catcher / spy-tag / dimer / TOXIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationcytolysis in another organism / The NLRP3 inflammasome / Purinergic signaling in leishmaniasis infection / toxin activity / extracellular region / identical protein binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
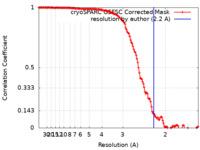
| Method | single particle reconstruction / cryo EM / Resolution: 2.2 Å | |||||||||
 Authors Authors | Ishii Y / Naito K / Yokoyama T / Tanaka Y / Matsuura T | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: alpha-Hemolysin(G122S/K147R)-SpyTag/SpyCatcher head to head 14-mer Authors: Ishii Y / Naito K / Yokoyama T / Tanaka Y / Matsuura T | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36688.map.gz emd_36688.map.gz | 126.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36688-v30.xml emd-36688-v30.xml emd-36688.xml emd-36688.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36688_fsc.xml emd_36688_fsc.xml | 14.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_36688.png emd_36688.png | 85 KB | ||
| Filedesc metadata |  emd-36688.cif.gz emd-36688.cif.gz | 5.6 KB | ||
| Others |  emd_36688_half_map_1.map.gz emd_36688_half_map_1.map.gz emd_36688_half_map_2.map.gz emd_36688_half_map_2.map.gz | 226.9 MB 226.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36688 http://ftp.pdbj.org/pub/emdb/structures/EMD-36688 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36688 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36688 | HTTPS FTP |
-Validation report
| Summary document |  emd_36688_validation.pdf.gz emd_36688_validation.pdf.gz | 915 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36688_full_validation.pdf.gz emd_36688_full_validation.pdf.gz | 914.6 KB | Display | |
| Data in XML |  emd_36688_validation.xml.gz emd_36688_validation.xml.gz | 22.6 KB | Display | |
| Data in CIF |  emd_36688_validation.cif.gz emd_36688_validation.cif.gz | 29.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36688 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36688 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36688 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36688 | HTTPS FTP |
-Related structure data
| Related structure data |  8jx2MC  8jx3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36688.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36688.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | B-factor Sharpened map with B-factor value of -60 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.788 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: one of the half maps
| File | emd_36688_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | one of the half maps | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: one of the half maps
| File | emd_36688_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | one of the half maps | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : heptameric alpha hemolysin pore dimerized by spy-catcher and spy-tag
| Entire | Name: heptameric alpha hemolysin pore dimerized by spy-catcher and spy-tag |
|---|---|
| Components |
|
-Supramolecule #1: heptameric alpha hemolysin pore dimerized by spy-catcher and spy-tag
| Supramolecule | Name: heptameric alpha hemolysin pore dimerized by spy-catcher and spy-tag type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: alpha hemolysin fused with spy-catcher
| Macromolecule | Name: alpha hemolysin fused with spy-catcher / type: protein_or_peptide / ID: 1 / Details: 300-412:SpyCatcher / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 47.003504 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MADSDINIKT GTTDIGSNTT VKTGDLVTYD KENGMHKKVF YSFIDDKNHN KKLLVIRTKG TIAGQYRVYS EEGANKSGLA WPSAFKVQL QLPDNEVAQI SDYYPRNSID TKEYMSTLTY GFNSNVTGDD TGKIGGLIGA NVSIGHTLRY VQPDFKTILE S PTDKKVGW ...String: MADSDINIKT GTTDIGSNTT VKTGDLVTYD KENGMHKKVF YSFIDDKNHN KKLLVIRTKG TIAGQYRVYS EEGANKSGLA WPSAFKVQL QLPDNEVAQI SDYYPRNSID TKEYMSTLTY GFNSNVTGDD TGKIGGLIGA NVSIGHTLRY VQPDFKTILE S PTDKKVGW KVIFNNMVNQ NWGPYDRDSW NPVYGNQLFM KTRNGSMKAA DNFLDPNKAS SLLSSGFSPD FATVITMDRK AS KQQTNID VIYERVRDDY QLHWTSTNWK GTNTKDKWTD RSSERYKIDW EKEEMTNGSS GSVTTLSGLS GEQGPSGDMT TEE DSATHI KFSKRDEDGR ELAGATMELR DSSGKTISTW ISDGHVKDFY LYPGKYTFVE TAAPDGYEVA TPIEFTVNED GQVT VDGEA TEGDAHTGGS HHHHHH UniProtKB: Alpha-hemolysin |
-Macromolecule #2: alpha hemolysin fused with spy-tag
| Macromolecule | Name: alpha hemolysin fused with spy-tag / type: protein_or_peptide / ID: 2 / Details: 300-315:SpyTag / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 36.803934 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MADSDINIKT GTTDIGSNTT VKTGDLVTYD KENGMHKKVF YSFIDDKNHN KKLLVIRTKG TIAGQYRVYS EEGANKSGLA WPSAFKVQL QLPDNEVAQI SDYYPRNSID TKEYMSTLTY GFNSNVTGDD TGKIGGLIGA NVSIGHTLRY VQPDFKTILE S PTDKKVGW ...String: MADSDINIKT GTTDIGSNTT VKTGDLVTYD KENGMHKKVF YSFIDDKNHN KKLLVIRTKG TIAGQYRVYS EEGANKSGLA WPSAFKVQL QLPDNEVAQI SDYYPRNSID TKEYMSTLTY GFNSNVTGDD TGKIGGLIGA NVSIGHTLRY VQPDFKTILE S PTDKKVGW KVIFNNMVNQ NWGPYDRDSW NPVYGNQLFM KTRNGSMKAA DNFLDPNKAS SLLSSGFSPD FATVITMDRK AS KQQTNID VIYERVRDDY QLHWTSTNWK GTNTKDKWTD RSSERYKIDW EKEEMTNGSS GSRGVPHIVM VDAYKRYKGG SHH HHHH UniProtKB: Alpha-hemolysin |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.58 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.6 Component:
Details: 20mM HEPES-KOH[pH7.6], 50mM Potassium glutamate | ||||||
| Grid | Model: Quantifoil R1.2/1.3 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE | ||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)