[English] 日本語
 Yorodumi
Yorodumi- EMDB-3596: The structure of the ESX-5 mycobacterial type VII secretion syste... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3596 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The structure of the ESX-5 mycobacterial type VII secretion system membrane complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Mycobacterium xenopi RIVM700367 (bacteria) Mycobacterium xenopi RIVM700367 (bacteria) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 13.1 Å | |||||||||
 Authors Authors | Ciccarelli L / Marlovits TC | |||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2017 Journal: Nat Microbiol / Year: 2017Title: Structure of the mycobacterial ESX-5 type VII secretion system membrane complex by single-particle analysis. Authors: Katherine S H Beckham / Luciano Ciccarelli / Catalin M Bunduc / Haydyn D T Mertens / Roy Ummels / Wolfgang Lugmayr / Julia Mayr / Mandy Rettel / Mikhail M Savitski / Dmitri I Svergun / ...Authors: Katherine S H Beckham / Luciano Ciccarelli / Catalin M Bunduc / Haydyn D T Mertens / Roy Ummels / Wolfgang Lugmayr / Julia Mayr / Mandy Rettel / Mikhail M Savitski / Dmitri I Svergun / Wilbert Bitter / Matthias Wilmanns / Thomas C Marlovits / Annabel H A Parret / Edith N G Houben /    Abstract: Mycobacteria are characterized by their impermeable outer membrane, which is rich in mycolic acids. To transport substrates across this complex cell envelope, mycobacteria rely on type VII (also ...Mycobacteria are characterized by their impermeable outer membrane, which is rich in mycolic acids. To transport substrates across this complex cell envelope, mycobacteria rely on type VII (also known as ESX) secretion systems. In Mycobacterium tuberculosis, these ESX systems are essential for growth and full virulence and therefore represent an attractive target for anti-tuberculosis drugs. However, the molecular details underlying type VII secretion are largely unknown, due to a lack of structural information. Here, we report the molecular architecture of the ESX-5 membrane complex from Mycobacterium xenopi determined at 13 Å resolution by electron microscopy. The four core proteins of the ESX-5 complex (EccB, EccC, EccD and EccE) assemble with equimolar stoichiometry into an oligomeric assembly that displays six-fold symmetry. This membrane-associated complex seems to be embedded exclusively in the inner membrane, which indicates that additional components are required to translocate substrates across the mycobacterial outer membrane. Furthermore, the extended cytosolic domains of the EccC ATPase, which interact with secretion effectors, are highly flexible, suggesting an as yet unseen mode of substrate interaction. Comparison of our results with known structures of other bacterial secretion systems demonstrates that the architecture of type VII secretion system is fundamentally different, suggesting an alternative secretion mechanism. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3596.map.gz emd_3596.map.gz | 9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3596-v30.xml emd-3596-v30.xml emd-3596.xml emd-3596.xml | 11.5 KB 11.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3596.jpg emd_3596.jpg emd_3596.png emd_3596.png | 141.8 KB 85.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3596 http://ftp.pdbj.org/pub/emdb/structures/EMD-3596 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3596 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3596 | HTTPS FTP |
-Validation report
| Summary document |  emd_3596_validation.pdf.gz emd_3596_validation.pdf.gz | 206.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3596_full_validation.pdf.gz emd_3596_full_validation.pdf.gz | 205.4 KB | Display | |
| Data in XML |  emd_3596_validation.xml.gz emd_3596_validation.xml.gz | 7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3596 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3596 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3596 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3596 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3596.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3596.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.18 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
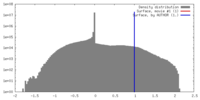
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : ESX-5 T7SS membrane complex
| Entire | Name: ESX-5 T7SS membrane complex |
|---|---|
| Components |
|
-Supramolecule #1: ESX-5 T7SS membrane complex
| Supramolecule | Name: ESX-5 T7SS membrane complex / type: complex / ID: 1 / Parent: 0 / Details: Solubilized in amphipol A8-35 |
|---|---|
| Source (natural) | Organism:  Mycobacterium xenopi RIVM700367 (bacteria) / Strain: RIVM700367 Mycobacterium xenopi RIVM700367 (bacteria) / Strain: RIVM700367 |
| Recombinant expression | Organism:  Mycobacterium smegmatis str. MC2 155 (bacteria) / Recombinant plasmid: pMV Mycobacterium smegmatis str. MC2 155 (bacteria) / Recombinant plasmid: pMV |
| Molecular weight | Theoretical: 1.8 MDa |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 20 mM Tris pH 8, 100 mM NaCl, 5 % (v/v) glycerol |
| Staining | Type: NEGATIVE / Material: Uranyl Acetate Details: A drop of 2% (w/v) uranyl acetate was added to a carbon-coated grid with absorbed protein and blotted after 30 s. |
| Grid | Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 5.0 nm / Pretreatment - Type: GLOW DISCHARGE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 20 |
|---|---|
| Image recording | Film or detector model: FEI EAGLE (4k x 4k) / Number grids imaged: 1 / Number real images: 65 / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source: LAB6 |
| Electron optics | Calibrated magnification: 44000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 1.2 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.4 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)