+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the PEA-bound mTAAR9 complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | PEA / mTAAR9 / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationAmine ligand-binding receptors / G alpha (s) signalling events / trace-amine receptor activity / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.17 Å | |||||||||
 Authors Authors | Sun JP / Li Q / Yang F / Xu YF / Guo LL / Lian S / Zhang MH / Rong NK | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Structural basis of amine odorant perception by a mammal olfactory receptor. Authors: Lulu Guo / Jie Cheng / Shuo Lian / Qun Liu / Yan Lu / Yuan Zheng / Kongkai Zhu / Minghui Zhang / Yalei Kong / Chao Zhang / Naikang Rong / Yuming Zhuang / Guoxing Fang / Jingjing Jiang / ...Authors: Lulu Guo / Jie Cheng / Shuo Lian / Qun Liu / Yan Lu / Yuan Zheng / Kongkai Zhu / Minghui Zhang / Yalei Kong / Chao Zhang / Naikang Rong / Yuming Zhuang / Guoxing Fang / Jingjing Jiang / Tianyao Zhang / Xiang Han / Zili Liu / Ming Xia / Shangming Liu / Lei Zhang / Stephen D Liberles / Xiao Yu / Yunfei Xu / Fan Yang / Qian Li / Jin-Peng Sun /   Abstract: Odorants are detected as smell in the nasal epithelium of mammals by two G-protein-coupled receptor families, the odorant receptors and the trace amine-associated receptors (TAARs). TAARs emerged ...Odorants are detected as smell in the nasal epithelium of mammals by two G-protein-coupled receptor families, the odorant receptors and the trace amine-associated receptors (TAARs). TAARs emerged following the divergence of jawed and jawless fish, and comprise a large monophyletic family of receptors that recognize volatile amine odorants to elicit both intraspecific and interspecific innate behaviours such as attraction and aversion. Here we report cryo-electron microscopy structures of mouse TAAR9 (mTAAR9) and mTAAR9-G or mTAAR9-G trimers in complex with β-phenylethylamine, N,N-dimethylcyclohexylamine or spermidine. The mTAAR9 structures contain a deep and tight ligand-binding pocket decorated with a conserved DWY motif, which is essential for amine odorant recognition. In the mTAAR9 structure, a unique disulfide bond connecting the N terminus to ECL2 is required for agonist-induced receptor activation. We identify key structural motifs of TAAR family members for detecting monoamines and polyamines and the shared sequence of different TAAR members that are responsible for recognition of the same odour chemical. We elucidate the molecular basis of mTAAR9 coupling to G and G by structural characterization and mutational analysis. Collectively, our results provide a structural basis for odorant detection, receptor activation and G coupling of an amine olfactory receptor. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35771.map.gz emd_35771.map.gz | 16.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35771-v30.xml emd-35771-v30.xml emd-35771.xml emd-35771.xml | 14.9 KB 14.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35771.png emd_35771.png | 62.4 KB | ||
| Filedesc metadata |  emd-35771.cif.gz emd-35771.cif.gz | 5.4 KB | ||
| Others |  emd_35771_half_map_1.map.gz emd_35771_half_map_1.map.gz emd_35771_half_map_2.map.gz emd_35771_half_map_2.map.gz | 17 MB 17 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35771 http://ftp.pdbj.org/pub/emdb/structures/EMD-35771 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35771 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35771 | HTTPS FTP |
-Validation report
| Summary document |  emd_35771_validation.pdf.gz emd_35771_validation.pdf.gz | 667.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35771_full_validation.pdf.gz emd_35771_full_validation.pdf.gz | 666.8 KB | Display | |
| Data in XML |  emd_35771_validation.xml.gz emd_35771_validation.xml.gz | 9.9 KB | Display | |
| Data in CIF |  emd_35771_validation.cif.gz emd_35771_validation.cif.gz | 11.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35771 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35771 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35771 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35771 | HTTPS FTP |
-Related structure data
| Related structure data |  8iwmMC  8itfC  8iw1C  8iw4C  8iw7C  8iw9C  8iweC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35771.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35771.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.076 Å | ||||||||||||||||||||||||||||||||||||
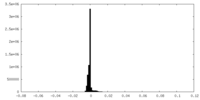
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_35771_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
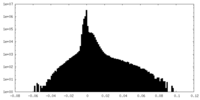
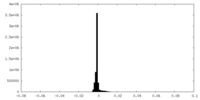
| Density Histograms |
-Half map: #2
| File | emd_35771_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
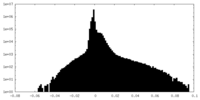
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of the PEA-bound mTAAR9 complex
| Entire | Name: Cryo-EM structure of the PEA-bound mTAAR9 complex |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of the PEA-bound mTAAR9 complex
| Supramolecule | Name: Cryo-EM structure of the PEA-bound mTAAR9 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Trace amine-associated receptor 9
| Macromolecule | Name: Trace amine-associated receptor 9 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 38.928238 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTSDFSPEPP MELCYENVNG SCIKSSYAPW PRAILYGVLG LGALLAVFGN LLVIIAILHF KQLHTPTNFL VASLACADFL VGVTVMPFS TVRSVESCWY FGESYCKFHT CFDTSFCFAS LFHLCCISID RYIAVTDPLT YPTKFTVSVS GLCIALSWFF S VTYSFSIF ...String: MTSDFSPEPP MELCYENVNG SCIKSSYAPW PRAILYGVLG LGALLAVFGN LLVIIAILHF KQLHTPTNFL VASLACADFL VGVTVMPFS TVRSVESCWY FGESYCKFHT CFDTSFCFAS LFHLCCISID RYIAVTDPLT YPTKFTVSVS GLCIALSWFF S VTYSFSIF YTGANEEGIE ELVVALTCVG GCQAPLNQNW VLLCFLLFFL PTVVMVFLYG RIFLVAKYQA RKIEGTANQA QA SSESYKE RVAKRERKAA KTLGIAMAAF LVSWLPYIID AVIDAYMNFI TPAYVYEILV WCVYYNSAMN PLIYAFFYPW FRK AIKLIV SGKVFRADSS TTNLFSEEAG AG UniProtKB: Trace amine-associated receptor 9 |
-Macromolecule #2: 2-PHENYLETHYLAMINE
| Macromolecule | Name: 2-PHENYLETHYLAMINE / type: ligand / ID: 2 / Number of copies: 1 / Formula: PEA |
|---|---|
| Molecular weight | Theoretical: 122.188 Da |
| Chemical component information |  ChemComp-PEA: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 1.875 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DIFFRACTION / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.17 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 463012 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)