+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the Mex67-Mtr2-3 heterodimer | |||||||||
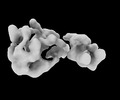
 Map data Map data | map of Mex67-Mtr2-3 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | nuclear export factor / NUCLEAR PROTEIN / RIBOSOME | |||||||||
| Biological species |  | |||||||||
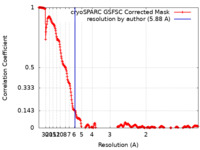
| Method | single particle reconstruction / cryo EM / Resolution: 5.88 Å | |||||||||
 Authors Authors | Li ZQ / Chen SJ / Sui SF | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Nuclear export of pre-60S particles through the nuclear pore complex. Authors: Zongqiang Li / Shuaijiabin Chen / Liang Zhao / Guoqiang Huang / Huiqin Xu / Xiaoyun Yang / Peiyi Wang / Ning Gao / Sen-Fang Sui /  Abstract: The nuclear pore complex (NPC) is the bidirectional gate that mediates the exchange of macromolecules or their assemblies between nucleus and cytoplasm. The assembly intermediates of the ribosomal ...The nuclear pore complex (NPC) is the bidirectional gate that mediates the exchange of macromolecules or their assemblies between nucleus and cytoplasm. The assembly intermediates of the ribosomal subunits, pre-60S and pre-40S particles, are among the largest cargoes of the NPC and the export of these gigantic ribonucleoproteins requires numerous export factors. Here we report the cryo-electron microscopy structure of native pre-60S particles trapped in the channel of yeast NPCs. In addition to known assembly factors, multiple factors with export functions are also included in the structure. These factors in general bind to either the flexible regions or subunit interface of the pre-60S particle, and virtually form many anchor sites for NPC binding. Through interactions with phenylalanine-glycine (FG) repeats from various nucleoporins of NPC, these factors collectively facilitate the passage of the pre-60S particle through the central FG repeat network of the NPC. Moreover, in silico analysis of the axial and radial distribution of pre-60S particles within the NPC shows that a single NPC can take up to four pre-60S particles simultaneously, and pre-60S particles are enriched in the inner ring regions close to the wall of the NPC with the solvent-exposed surface facing the centre of the nuclear pore. Our data suggest a translocation model for the export of pre-60S particles through the NPC. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35767.map.gz emd_35767.map.gz | 950.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35767-v30.xml emd-35767-v30.xml emd-35767.xml emd-35767.xml | 13.3 KB 13.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35767_fsc.xml emd_35767_fsc.xml | 26.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_35767.png emd_35767.png | 33.8 KB | ||
| Others |  emd_35767_half_map_1.map.gz emd_35767_half_map_1.map.gz emd_35767_half_map_2.map.gz emd_35767_half_map_2.map.gz | 1.8 GB 1.8 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35767 http://ftp.pdbj.org/pub/emdb/structures/EMD-35767 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35767 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35767 | HTTPS FTP |
-Validation report
| Summary document |  emd_35767_validation.pdf.gz emd_35767_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35767_full_validation.pdf.gz emd_35767_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_35767_validation.xml.gz emd_35767_validation.xml.gz | 37 KB | Display | |
| Data in CIF |  emd_35767_validation.cif.gz emd_35767_validation.cif.gz | 48.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35767 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35767 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35767 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35767 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_35767.map.gz / Format: CCP4 / Size: 1.9 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35767.map.gz / Format: CCP4 / Size: 1.9 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map of Mex67-Mtr2-3 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.668 Å | ||||||||||||||||||||||||||||||||||||
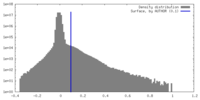
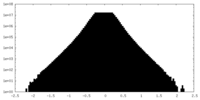
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half A map of Mex67-Mtr2-3
| File | emd_35767_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half A map of Mex67-Mtr2-3 | ||||||||||||
| Projections & Slices |
| ||||||||||||
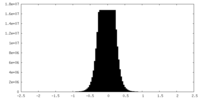
| Density Histograms |
-Half map: half B map of Mex67-Mtr2-3
| File | emd_35767_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half B map of Mex67-Mtr2-3 | ||||||||||||
| Projections & Slices |
| ||||||||||||
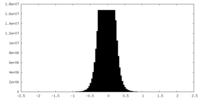
| Density Histograms |
- Sample components
Sample components
-Entire : Mex67-Mtr2 heterodimer
| Entire | Name: Mex67-Mtr2 heterodimer |
|---|---|
| Components |
|
-Supramolecule #1: Mex67-Mtr2 heterodimer
| Supramolecule | Name: Mex67-Mtr2 heterodimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Nuclear export factor
| Macromolecule | Name: Nuclear export factor / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MSGFHNVGNI NMMAQQQMQQ NRIKISVRNW QNATMNDLIN FISRNARVAV YDAHVEGPLV IGYVNSKAE AESLMKWNGV RFAGSNLKFE LLDNNGASAG TSDTISFLRG VLLKRYDPQT K LLNLGALH SDPELIQKGV FSSISTQSKM FPAMMKLAST EKSLIVESVN ...String: MSGFHNVGNI NMMAQQQMQQ NRIKISVRNW QNATMNDLIN FISRNARVAV YDAHVEGPLV IGYVNSKAE AESLMKWNGV RFAGSNLKFE LLDNNGASAG TSDTISFLRG VLLKRYDPQT K LLNLGALH SDPELIQKGV FSSISTQSKM FPAMMKLAST EKSLIVESVN LADNQLKDIS AI STLAQTF PNLKNLCLAN NQIFRFRSLE VWKNKFKDLR ELLMTNNPIT TDKLYRTEML RLF PKLVVL DNVIVRDEQK LQTVYSLPMK IQQFFFENDA LGQSSTDFAT NFLNLWDNNR EQLL NLYSP QSQFSVSVDS TIPPSTVTDS DQTPAFGYYM SSSRNISKVS SEKSIQQRLS IGQES INSI FKTLPKTKHH LQEQPNEYSM ETISYPQING FVITLHGFFE ETGKPELESN KKTGKN NYQ KNRRYNHGYN STSNNKLSKK SFDRTWVIVP MNNSVIIASD LLTVRAYSTG AWKTASI AI AQPPQQQASV LPQVASMNPN ITTPPQPQPS VVPGGMSIPG APQGAMVMAP TLQLPPDV Q SRLNPVQLEL LNKLHLETKL NAEYTFMLAE QSNWNYEVAI KGFQSSMNGI PREAFVQF |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)