+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3542 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of a spliceosome remodeled for exon ligation | |||||||||
 Map data Map data | Map of Saccharomyces cerevisiae spliceosome C-star complex generated by focussed refinement on U2 snRNP peripheral regions. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   | |||||||||
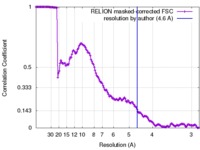
| Method | single particle reconstruction / cryo EM / Resolution: 4.6 Å | |||||||||
 Authors Authors | Fica SM / Oubridge C / Galej WP / Wilkinson ME / Newman AJ / Bai X-C / Nagai K | |||||||||
 Citation Citation |  Journal: Nature / Year: 2016 Journal: Nature / Year: 2016Title: Cryo-EM structure of the spliceosome immediately after branching. Authors: Wojciech P Galej / Max E Wilkinson / Sebastian M Fica / Chris Oubridge / Andrew J Newman / Kiyoshi Nagai /  Abstract: Precursor mRNA (pre-mRNA) splicing proceeds by two consecutive transesterification reactions via a lariat-intron intermediate. Here we present the 3.8 Å cryo-electron microscopy structure of the ...Precursor mRNA (pre-mRNA) splicing proceeds by two consecutive transesterification reactions via a lariat-intron intermediate. Here we present the 3.8 Å cryo-electron microscopy structure of the spliceosome immediately after lariat formation. The 5'-splice site is cleaved but remains close to the catalytic Mg site in the U2/U6 small nuclear RNA (snRNA) triplex, and the 5'-phosphate of the intron nucleotide G(+1) is linked to the branch adenosine 2'OH. The 5'-exon is held between the Prp8 amino-terminal and linker domains, and base-pairs with U5 snRNA loop 1. Non-Watson-Crick interactions between the branch helix and 5'-splice site dock the branch adenosine into the active site, while intron nucleotides +3 to +6 base-pair with the U6 snRNA ACAGAGA sequence. Isy1 and the step-one factors Yju2 and Cwc25 stabilize docking of the branch helix. The intron downstream of the branch site emerges between the Prp8 reverse transcriptase and linker domains and extends towards the Prp16 helicase, suggesting a plausible mechanism of remodelling before exon ligation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3542.map.gz emd_3542.map.gz | 243.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3542-v30.xml emd-3542-v30.xml emd-3542.xml emd-3542.xml | 59.3 KB 59.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3542_fsc.xml emd_3542_fsc.xml | 14.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_3542.png emd_3542.png | 62.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3542 http://ftp.pdbj.org/pub/emdb/structures/EMD-3542 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3542 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3542 | HTTPS FTP |
-Validation report
| Summary document |  emd_3542_validation.pdf.gz emd_3542_validation.pdf.gz | 312.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3542_full_validation.pdf.gz emd_3542_full_validation.pdf.gz | 311.5 KB | Display | |
| Data in XML |  emd_3542_validation.xml.gz emd_3542_validation.xml.gz | 14 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3542 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3542 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3542 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3542 | HTTPS FTP |
-Related structure data
| Related structure data |  3539C  3541C  5mpsC  5mq0C C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10687 (Title: Yeast C, Ci, C*, and P complex spliceosomes / Data size: 8.9 TB EMPIAR-10687 (Title: Yeast C, Ci, C*, and P complex spliceosomes / Data size: 8.9 TBData #1: Unaligned movies of C-complex spliceosome with 3' splice site AG to AC mutation (Dataset 1) [micrographs - multiframe] Data #2: Unaligned movies of C and C*-complex spliceosomes with 3' splice site AG to AdG mutation (Dataset 2) [micrographs - multiframe] Data #3: Unaligned movies of C and C*-complex spliceosomes with 3' splice site AG to AdG mutation (Dataset 3) [micrographs - multiframe] Data #4: Aligned movies of C-complex spliceosomes with cold-sensitive prp16-302 mutation, purified with Cwc25 (Dataset 4) [micrographs - multiframe] Data #5: Unaligned movies of C-complex spliceosomes with cold-sensitive prp16-302 mutation, purified with Cwc25 and incubated with ATP and Mg (Dataset 5) [micrographs - multiframe] Data #6: Unaligned movies of C, C*, and P-complex spliceosomes with dominant-negative Prp22 mutation K512A, purified with Slu7 (Dataset 6) [micrographs - multiframe] Data #7: Unaligned movies of P-complex spliceosomes with dominant-negative Prp22 mutation K512A, treated with anti-3'exon RNaseH oligo, purified in presence of Mg (Dataset 9) [micrographs - single frame] Data #8: Selected C-complex particles after polishing [picked particles - single frame - processed] Data #9: Selected P-complex particles after polishing [picked particles - single frame - processed] Data #10: Various signal subtractions for C- and P-complex spliceosomes [picked particles - single frame - processed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3542.map.gz / Format: CCP4 / Size: 266.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3542.map.gz / Format: CCP4 / Size: 266.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map of Saccharomyces cerevisiae spliceosome C-star complex generated by focussed refinement on U2 snRNP peripheral regions. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.43 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Saccharomyces cerevisiae spliceosome. Complex C just after Prp16-...
+Supramolecule #1: Saccharomyces cerevisiae spliceosome. Complex C just after Prp16-...
+Macromolecule #1: Intron of UBC4 pre-mRNA
+Macromolecule #2: 5'exon of UBC4 pre-mRNA
+Macromolecule #3: U2 snRNA
+Macromolecule #4: U6 snRNA
+Macromolecule #5: U5 snRNA
+Macromolecule #34: 3'exon of UBC4 pre-mRNA bound to Prp22 helicase
+Macromolecule #6: Pre-mRNA-splicing factor 8
+Macromolecule #7: Pre-mRNA-splicing factor SNU114
+Macromolecule #8: Pre-mRNA-splicing factor CWC22
+Macromolecule #9: Pre-mRNA-splicing factor PRP46
+Macromolecule #10: Pre-mRNA-processing protein 45
+Macromolecule #11: Pre-mRNA-splicing factor BUD31
+Macromolecule #12: Pre-mRNA-splicing factor CEF1
+Macromolecule #13: Pre-mRNA-splicing factor CWC15
+Macromolecule #14: Pre-mRNA-splicing factor CWC21
+Macromolecule #15: Pre-mRNA-splicing factor CLF1
+Macromolecule #16: Pre-mRNA-splicing factor SYF1
+Macromolecule #17: Pre-mRNA-splicing factor 18
+Macromolecule #18: Pre-mRNA-splicing factor SLU7
+Macromolecule #19: Pre-mRNA-splicing factor SYF2
+Macromolecule #20: Small nuclear ribonucleoprotein-associated protein B
+Macromolecule #21: Small nuclear ribonucleoprotein Sm D3
+Macromolecule #22: Small nuclear ribonucleoprotein E
+Macromolecule #23: Small nuclear ribonucleoprotein F
+Macromolecule #24: Small nuclear ribonucleoprotein G
+Macromolecule #25: Small nuclear ribonucleoprotein Sm D1
+Macromolecule #26: Small nuclear ribonucleoprotein Sm D2
+Macromolecule #27: Pre-mRNA-splicing factor CWC2
+Macromolecule #28: Pre-mRNA-splicing factor SLT11
+Macromolecule #29: Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP22
+Macromolecule #30: Pre-mRNA-processing factor 17
+Macromolecule #31: U2 small nuclear ribonucleoprotein A'
+Macromolecule #32: U2 small nuclear ribonucleoprotein B''
+Macromolecule #33: Unknown protein
+Macromolecule #35: MAGNESIUM ION
+Macromolecule #36: POTASSIUM ION
+Macromolecule #37: INOSITOL HEXAKISPHOSPHATE
+Macromolecule #38: GUANOSINE-5'-TRIPHOSPHATE
+Macromolecule #39: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.9 Component:
Details: NP-40 is also called IGEPAL CA-630 | ||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Support film - Film thickness: 6.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 20.0 kPa | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK III Details: 3.5 microlitres sample were applied to the grid, left for 25 seconds and then blotted for 3.0-3.5 seconds before plunging.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum energy filter, 20 eV slit width |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number real images: 3596 / Average exposure time: 0.8 sec. / Average electron dose: 2.0 e/Å2 Details: Total dose: 40 electrons/Angstrom^2 over 16 seconds. 20 movie frames collected at 1.25 frames per second. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | Used secondary structure restraints generated in ProSMART and LibG for Refmac refinement. |
|---|---|
| Refinement | Space: RECIPROCAL / Protocol: FLEXIBLE FIT / Overall B value: 330 / Target criteria: Fourier Shell Correlation |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)