[English] 日本語
 Yorodumi
Yorodumi- EMDB-3527: S. pombe microtubule decorated with Cut7 motor domain in the AMPP... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3527 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
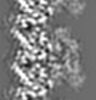
| Title | S. pombe microtubule decorated with Cut7 motor domain in the AMPPNP state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | S. pombe / microtubule / kinesin-5 / Cut7 motor domain / motor protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationmitotic spindle pole body duplication / Platelet degranulation / mitotic spindle formation (spindle phase one) / mitotic spindle elongation (spindle phase three) / Kinesins / initial mitotic spindle pole body separation / microtubule plus-end directed mitotic chromosome migration / meiotic spindle assembly / nuclear migration by microtubule mediated pushing forces / meiotic spindle pole ...mitotic spindle pole body duplication / Platelet degranulation / mitotic spindle formation (spindle phase one) / mitotic spindle elongation (spindle phase three) / Kinesins / initial mitotic spindle pole body separation / microtubule plus-end directed mitotic chromosome migration / meiotic spindle assembly / nuclear migration by microtubule mediated pushing forces / meiotic spindle pole / mitotic spindle elongation / nuclear division / mitotic spindle midzone / mitotic spindle midzone assembly / mitotic spindle pole body / spindle elongation / astral microtubule / polar microtubule / minus-end-directed microtubule motor activity / plus-end-directed microtubule motor activity / meiotic spindle / microtubule associated complex / microtubule motor activity / intracellular distribution of mitochondria / cytoplasmic microtubule / mitotic spindle assembly / cytoplasmic microtubule organization / spindle microtubule / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / mitotic spindle / structural constituent of cytoskeleton / kinetochore / spindle / microtubule cytoskeleton organization / microtubule cytoskeleton / mitotic cell cycle / microtubule binding / microtubule / hydrolase activity / cell division / response to antibiotic / GTPase activity / GTP binding / ATP hydrolysis activity / ATP binding / nucleus / metal ion binding / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.5 Å | |||||||||
 Authors Authors | Moores CA / von Loeffelholz O | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation | Journal: J Mol Biol / Year: 2020 Title: Corrigendum to "Cryo-EM Structure (4.5 Å) of Yeast Kinesin-5-Microtubule Complex Reveals a Distinct Binding Footprint and Mechanism of Drug Resistance" [J. Mol. Biol. 431 (2019) 864-872] ...Title: Corrigendum to "Cryo-EM Structure (4.5 Å) of Yeast Kinesin-5-Microtubule Complex Reveals a Distinct Binding Footprint and Mechanism of Drug Resistance" [J. Mol. Biol. 431 (2019) 864-872] https://doi.org/10.1016/j.jmb.2019.01.011. Authors: Ottilie von Loeffelholz / Alejandro Peña / Douglas Robert Drummond / Robert Cross / Carolyn Ann Moores /    | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3527.map.gz emd_3527.map.gz | 5.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3527-v30.xml emd-3527-v30.xml emd-3527.xml emd-3527.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3527.png emd_3527.png | 143.7 KB | ||
| Others |  emd_3527_additional.map.gz emd_3527_additional.map.gz emd_3527_additional_1.map.gz emd_3527_additional_1.map.gz | 1.2 MB 1.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3527 http://ftp.pdbj.org/pub/emdb/structures/EMD-3527 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3527 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3527 | HTTPS FTP |
-Validation report
| Summary document |  emd_3527_validation.pdf.gz emd_3527_validation.pdf.gz | 526 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3527_full_validation.pdf.gz emd_3527_full_validation.pdf.gz | 525.6 KB | Display | |
| Data in XML |  emd_3527_validation.xml.gz emd_3527_validation.xml.gz | 4.4 KB | Display | |
| Data in CIF |  emd_3527_validation.cif.gz emd_3527_validation.cif.gz | 4.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3527 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3527 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3527 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3527 | HTTPS FTP |
-Related structure data
| Related structure data |  6s8mMC  5mlv M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3527.map.gz / Format: CCP4 / Size: 1.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3527.map.gz / Format: CCP4 / Size: 1.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.39 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
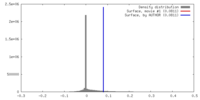
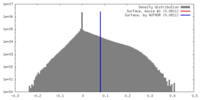
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: #1
| File | emd_3527_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
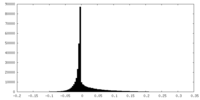
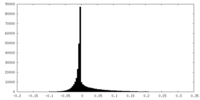
| Density Histograms |
-Additional map: #1
| File | emd_3527_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
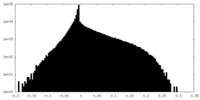
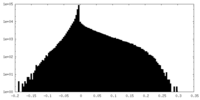
| Density Histograms |
- Sample components
Sample components
+Entire : Cut7 MTD decorated S. pombe microtubule stabilized with Epothilone-B
+Supramolecule #1: Cut7 MTD decorated S. pombe microtubule stabilized with Epothilone-B
+Supramolecule #2: Alpha tubulin, Beta tubulin
+Supramolecule #3: Cut7
+Macromolecule #1: Tubulin alpha-1 chain
+Macromolecule #2: Kinesin-like protein cut7
+Macromolecule #3: Tubulin beta chain
+Macromolecule #4: GUANOSINE-5'-TRIPHOSPHATE
+Macromolecule #5: MAGNESIUM ION
+Macromolecule #6: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER
+Macromolecule #7: GUANOSINE-5'-DIPHOSPHATE
+Macromolecule #8: 7,11-DIHYDROXY-8,8,10,12,16-PENTAMETHYL-3-[1-METHYL-2-(2-METHYL-T...
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 6.5 |
|---|---|
| Sugar embedding | Material: vitreous ice |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.5 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: FREALIGN Details: reference was low-pass filtered to 8 Angstroms in the final round of alignment. Number images used: 12543 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)