[English] 日本語
 Yorodumi
Yorodumi- EMDB-35010: Human Consensus Olfactory Receptor OR52c in Complex with Octanoic... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human Consensus Olfactory Receptor OR52c in Complex with Octanoic Acid (OCA) and G Protein | |||||||||
 Map data Map data | This is a map of Human consensus olfactory receptor OR52c in complex with octanoic acid and G protein. It is a merged map of receptor-focused and G protein-focused local refinement maps. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Olfactory Receptor / G Protein / MEMBRANE PROTEIN / GPCR / Olfactory GPCR | |||||||||
| Function / homology |  Function and homology information Function and homology informationPKA activation in glucagon signalling / hair follicle placode formation / developmental growth / D1 dopamine receptor binding / intracellular transport / renal water homeostasis / Hedgehog 'off' state / adenylate cyclase-activating adrenergic receptor signaling pathway / activation of adenylate cyclase activity / cellular response to glucagon stimulus ...PKA activation in glucagon signalling / hair follicle placode formation / developmental growth / D1 dopamine receptor binding / intracellular transport / renal water homeostasis / Hedgehog 'off' state / adenylate cyclase-activating adrenergic receptor signaling pathway / activation of adenylate cyclase activity / cellular response to glucagon stimulus / adenylate cyclase activator activity / regulation of insulin secretion / trans-Golgi network membrane / negative regulation of inflammatory response to antigenic stimulus / bone development / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / Activation of the phototransduction cascade / adenylate cyclase-activating G protein-coupled receptor signaling pathway / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / G protein activity / G-protein activation / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / platelet aggregation / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / cognition / G beta:gamma signalling through BTK / ADP signalling through P2Y purinoceptor 12 / Sensory perception of sweet, bitter, and umami (glutamate) taste / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / photoreceptor disc membrane / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / ADORA2B mediated anti-inflammatory cytokines production / cellular response to catecholamine stimulus / sensory perception of taste / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / cellular response to prostaglandin E stimulus / G-protein beta-subunit binding / Inactivation, recovery and regulation of the phototransduction cascade / heterotrimeric G-protein complex / sensory perception of smell / G alpha (12/13) signalling events / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / GTPase binding / positive regulation of cold-induced thermogenesis / retina development in camera-type eye / Ca2+ pathway / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (i) signalling events / fibroblast proliferation / G alpha (s) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / G alpha (q) signalling events / Ras protein signal transduction / cell population proliferation / Extra-nuclear estrogen signaling / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / synapse / protein-containing complex binding / GTP binding / signal transduction / extracellular exosome / membrane / metal ion binding / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.97 Å | |||||||||
 Authors Authors | Choi CW / Bae J / Choi H-J / Kim J | |||||||||
| Funding support |  Korea, Republic Of, 1 items Korea, Republic Of, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Understanding the molecular mechanisms of odorant binding and activation of the human OR52 family. Authors: Chulwon Choi / Jungnam Bae / Seonghan Kim / Seho Lee / Hyunook Kang / Jinuk Kim / Injin Bang / Kiheon Kim / Won-Ki Huh / Chaok Seok / Hahnbeom Park / Wonpil Im / Hee-Jung Choi /   Abstract: Structural and mechanistic studies on human odorant receptors (ORs), key in olfactory signaling, are challenging because of their low surface expression in heterologous cells. The recent structure of ...Structural and mechanistic studies on human odorant receptors (ORs), key in olfactory signaling, are challenging because of their low surface expression in heterologous cells. The recent structure of OR51E2 bound to propionate provided molecular insight into odorant recognition, but the lack of an inactive OR structure limited understanding of the activation mechanism of ORs upon odorant binding. Here, we determined the cryo-electron microscopy structures of consensus OR52 (OR52), a representative of the OR52 family, in the ligand-free (apo) and octanoate-bound states. The apo structure of OR52 reveals a large opening between transmembrane helices (TMs) 5 and 6. A comparison between the apo and active structures of OR52 demonstrates the inward and outward movements of the extracellular and intracellular segments of TM6, respectively. These results, combined with molecular dynamics simulations and signaling assays, shed light on the molecular mechanisms of odorant binding and activation of the OR52 family. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35010.map.gz emd_35010.map.gz | 228.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35010-v30.xml emd-35010-v30.xml emd-35010.xml emd-35010.xml | 28.5 KB 28.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35010_fsc.xml emd_35010_fsc.xml | 13.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_35010.png emd_35010.png | 128.9 KB | ||
| Filedesc metadata |  emd-35010.cif.gz emd-35010.cif.gz | 7.4 KB | ||
| Others |  emd_35010_additional_1.map.gz emd_35010_additional_1.map.gz emd_35010_additional_2.map.gz emd_35010_additional_2.map.gz emd_35010_half_map_1.map.gz emd_35010_half_map_1.map.gz emd_35010_half_map_2.map.gz emd_35010_half_map_2.map.gz | 230 MB 230 MB 226.5 MB 226.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35010 http://ftp.pdbj.org/pub/emdb/structures/EMD-35010 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35010 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35010 | HTTPS FTP |
-Validation report
| Summary document |  emd_35010_validation.pdf.gz emd_35010_validation.pdf.gz | 954.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35010_full_validation.pdf.gz emd_35010_full_validation.pdf.gz | 954 KB | Display | |
| Data in XML |  emd_35010_validation.xml.gz emd_35010_validation.xml.gz | 22 KB | Display | |
| Data in CIF |  emd_35010_validation.cif.gz emd_35010_validation.cif.gz | 28.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35010 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35010 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35010 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35010 | HTTPS FTP |
-Related structure data
| Related structure data |  8htiMC  8htgC  8j46C  8w77C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35010.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35010.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is a map of Human consensus olfactory receptor OR52c in complex with octanoic acid and G protein. It is a merged map of receptor-focused and G protein-focused local refinement maps. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.851 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: This is a map of Human consensus olfactory...
| File | emd_35010_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is a map of Human consensus olfactory receptor OR52c in complex with octanoic acid and G protein. It is a receptor-focused local refinement sharp map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: This is a map of Human consensus olfactory...
| File | emd_35010_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is a map of Human consensus olfactory receptor OR52c in complex with octanoic acid and G protein. It is a receptor-focused local refinement sharp map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: This is a map of Human consensus olfactory...
| File | emd_35010_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is a map of Human consensus olfactory receptor OR52c in complex with octanoic acid and G protein. It is a half B map of NU-refinement map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: This is a map of Human consensus olfactory...
| File | emd_35010_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is a map of Human consensus olfactory receptor OR52c in complex with octanoic acid and G protein. It is a half A map of NU-refinement map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of Consensus Olfactory receptor OR52c in complex with Oct...
| Entire | Name: Complex of Consensus Olfactory receptor OR52c in complex with Octanoic acid, G protein and Nanobody 35 (Nb35) |
|---|---|
| Components |
|
-Supramolecule #1: Complex of Consensus Olfactory receptor OR52c in complex with Oct...
| Supramolecule | Name: Complex of Consensus Olfactory receptor OR52c in complex with Octanoic acid, G protein and Nanobody 35 (Nb35) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 140 KDa |
-Macromolecule #1: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
| Macromolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 45.891809 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HHHHHHLEVL FQGPGSLGNS KTEDQRNEEK AQREANKKIE KQLQKDKQVY RATHRLLLLG AGESGKSTIV KQMRILHVNG FNGDSEKAT KVQDIKNNLK EAIETIVAAM SNLVPPVELA NPENQFRVDY ILSVMNVPDF DFPPEFYEHA KALWEDEGVR A CYERSNEY ...String: HHHHHHLEVL FQGPGSLGNS KTEDQRNEEK AQREANKKIE KQLQKDKQVY RATHRLLLLG AGESGKSTIV KQMRILHVNG FNGDSEKAT KVQDIKNNLK EAIETIVAAM SNLVPPVELA NPENQFRVDY ILSVMNVPDF DFPPEFYEHA KALWEDEGVR A CYERSNEY QLIDCAQYFL DKIDVIKQAD YVPSDQDLLR CRVLTSGIFE TKFQVDKVNF HMFDVGGQRD ERRKWIQCFN DV TAIIFVV ASSSYNMVIR EDNQTNRLQE ALNLFKSIWN NRWLRTISVI LFLNKQDLLA EKVLAGKSKI EDYFPEFARY TTP EDATPE PGEDPRVTRA KYFIRDEFLR ISTASGDGRH YCYPHFTCAV DTENIRRVFN DCRDIIQRMH LRQYELL UniProtKB: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.286891 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HHHHHHLEVL FQGPGSSGSE LDQLRQEAEQ LKNQIRDARK ACADATLSQI TNNIDPVGRI QMRTRRTLRG HLAKIYAMHW GTDSRLLVS ASQDGKLIIW DSYTTNKVHA IPLRSSWVMT CAYAPSGNYV ACGGLDNICS IYNLKTREGN VRVSRELAGH T GYLSCCRF ...String: HHHHHHLEVL FQGPGSSGSE LDQLRQEAEQ LKNQIRDARK ACADATLSQI TNNIDPVGRI QMRTRRTLRG HLAKIYAMHW GTDSRLLVS ASQDGKLIIW DSYTTNKVHA IPLRSSWVMT CAYAPSGNYV ACGGLDNICS IYNLKTREGN VRVSRELAGH T GYLSCCRF LDDNQIVTSS GDTTCALWDI ETGQQTTTFT GHTGDVMSLS LAPDTRLFVS GACDASAKLW DVREGMCRQT FT GHESDIN AICFFPNGNA FATGSDDATC RLFDLRADQE LMTYSHDNII CGITSVSFSK SGRLLLAGYD DFNCNVWDAL KAD RAGVLA GHDNRVSCLG VTDDGMAVAT GSWDSFLKIW N UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Nanobody 35
| Macromolecule | Name: Nanobody 35 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.140742 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLQESGGG LVQPGGSLRL SCAASGFTFS NYKMNWVRQA PGKGLEWVSD ISQSGASISY TGSVKGRFTI SRDNAKNTLY LQMNSLKPE DTAVYYCARC PAPFTRDCFD VTSTTYAYRG QGTQVTVSSH HHHHHEPEA |
-Macromolecule #5: Consensus Olfactory Receptor OR52c
| Macromolecule | Name: Consensus Olfactory Receptor OR52c / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 36.49775 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DYKDDDDAID MPTSNHTSFH PSSFLLVGIP GLESVHIWIS IPFCAMYLIA LLGNSTLLFV IKTERSLHEP MYYFLAMLAA TDLVLSTST IPKMLAIFWF NLKEISFDAC LTQMFFIHSF TGMESGVLLA MAFDRYVAIC YPLRYTTILT NKVIGKIGMA V VLRAVLLV ...String: DYKDDDDAID MPTSNHTSFH PSSFLLVGIP GLESVHIWIS IPFCAMYLIA LLGNSTLLFV IKTERSLHEP MYYFLAMLAA TDLVLSTST IPKMLAIFWF NLKEISFDAC LTQMFFIHSF TGMESGVLLA MAFDRYVAIC YPLRYTTILT NKVIGKIGMA V VLRAVLLV IPFPFLLKRL PFCGTNIIPH TYCEHMGVAK LACADIKVNI IYGLFVALLI VGLDVILIAL SYVLILRAVF RL PSQDARL KALSTCGSHI CVILAFYTPA FFSFLTHRFG HHIPPYIHIL LANLYLLVPP MLNPIIYGVK TKQIRERVLK IFF KKK |
-Macromolecule #6: OCTANOIC ACID (CAPRYLIC ACID)
| Macromolecule | Name: OCTANOIC ACID (CAPRYLIC ACID) / type: ligand / ID: 6 / Number of copies: 1 / Formula: OCA |
|---|---|
| Molecular weight | Theoretical: 144.211 Da |
| Chemical component information | 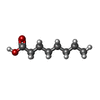 ChemComp-OCA: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 11 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV / Details: blot time 3 seconds. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-8hti: |
 Movie
Movie Controller
Controller



























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)