+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
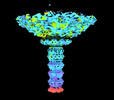
| Title | Dinoroseobacter phage vB_DshS-R4C C1 portal vertex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample | vB_DshS-R4C C1 portal vertex != Dinoroseobacter phage vB_DshS-R4C vB_DshS-R4C C1 portal vertex
| |||||||||
 Keywords Keywords | Marine bacteriophage / Cryo-EM / Portal vertex / VIRUS | |||||||||
| Biological species |  Dinoroseobacter phage vB_DshS-R4C (virus) Dinoroseobacter phage vB_DshS-R4C (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 11.5 Å | |||||||||
 Authors Authors | Huang Y / Sun H / Wei S / Zheng Q / Li S / Zhang R / Xia N | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structure and proposed DNA delivery mechanism of a marine roseophage. Authors: Yang Huang / Hui Sun / Shuzhen Wei / Lanlan Cai / Liqin Liu / Yanan Jiang / Jiabao Xin / Zhenqin Chen / Yuqiong Que / Zhibo Kong / Tingting Li / Hai Yu / Jun Zhang / Ying Gu / Qingbing Zheng ...Authors: Yang Huang / Hui Sun / Shuzhen Wei / Lanlan Cai / Liqin Liu / Yanan Jiang / Jiabao Xin / Zhenqin Chen / Yuqiong Que / Zhibo Kong / Tingting Li / Hai Yu / Jun Zhang / Ying Gu / Qingbing Zheng / Shaowei Li / Rui Zhang / Ningshao Xia /  Abstract: Tailed bacteriophages (order, Caudovirales) account for the majority of all phages. However, the long flexible tail of siphophages hinders comprehensive investigation of the mechanism of viral gene ...Tailed bacteriophages (order, Caudovirales) account for the majority of all phages. However, the long flexible tail of siphophages hinders comprehensive investigation of the mechanism of viral gene delivery. Here, we report the atomic capsid and in-situ structures of the tail machine of the marine siphophage, vB_DshS-R4C (R4C), which infects Roseobacter. The R4C virion, comprising 12 distinct structural protein components, has a unique five-fold vertex of the icosahedral capsid that allows genome delivery. The specific position and interaction pattern of the tail tube proteins determine the atypical long rigid tail of R4C, and further provide negative charge distribution within the tail tube. A ratchet mechanism assists in DNA transmission, which is initiated by an absorption device that structurally resembles the phage-like particle, RcGTA. Overall, these results provide in-depth knowledge into the intact structure and underlining DNA delivery mechanism for the ecologically important siphophages. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34253.map.gz emd_34253.map.gz | 192.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34253-v30.xml emd-34253-v30.xml emd-34253.xml emd-34253.xml | 12.8 KB 12.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34253.png emd_34253.png | 74.4 KB | ||
| Others |  emd_34253_half_map_1.map.gz emd_34253_half_map_1.map.gz emd_34253_half_map_2.map.gz emd_34253_half_map_2.map.gz | 193.8 MB 193.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34253 http://ftp.pdbj.org/pub/emdb/structures/EMD-34253 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34253 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34253 | HTTPS FTP |
-Validation report
| Summary document |  emd_34253_validation.pdf.gz emd_34253_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34253_full_validation.pdf.gz emd_34253_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_34253_validation.xml.gz emd_34253_validation.xml.gz | 15.9 KB | Display | |
| Data in CIF |  emd_34253_validation.cif.gz emd_34253_validation.cif.gz | 18.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34253 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34253 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34253 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34253 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34253.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34253.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.128 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_34253_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
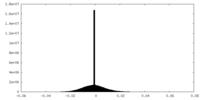
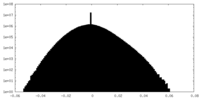
| Density Histograms |
-Half map: #2
| File | emd_34253_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
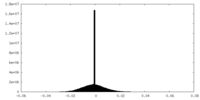
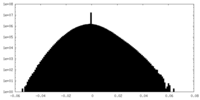
| Density Histograms |
- Sample components
Sample components
-Entire : vB_DshS-R4C C1 portal vertex
| Entire | Name: vB_DshS-R4C C1 portal vertex |
|---|---|
| Components |
|
-Supramolecule #1: Dinoroseobacter phage vB_DshS-R4C
| Supramolecule | Name: Dinoroseobacter phage vB_DshS-R4C / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 / NCBI-ID: 2590919 / Sci species name: Dinoroseobacter phage vB_DshS-R4C / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Dinoroseobacter shibae DFL 12 = DSM 16493 (bacteria) Dinoroseobacter shibae DFL 12 = DSM 16493 (bacteria) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER / Details: trRosetta server |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 11.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 5844 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)