[English] 日本語
 Yorodumi
Yorodumi- EMDB-34249: Cryo-EM model of the marine siphophage vB_DshS-R4C baseplate-tail... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
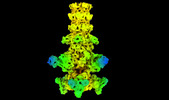
| Title | Cryo-EM model of the marine siphophage vB_DshS-R4C baseplate-tail complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Marine bacteriophage / Cryo-EM / Siphophage / Baseplate / Megatron protein / Tail fibre protein / Distal tail protein / Hub protein / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationProtein of unknown function DUF2793 / Protein of unknown function (DUF2793) / GTA TIM-barrel-like domain / : / GTA TIM-barrel-like domain / Rcc01698-like, C-terminal / Protein of unknown function DUF2460 / Conserved hypothetical protein 2217 (DUF2460) / Baseplate hub protein, N-terminal attachment domain / Bacteriophage phiJL001, Gp84 ...Protein of unknown function DUF2793 / Protein of unknown function (DUF2793) / GTA TIM-barrel-like domain / : / GTA TIM-barrel-like domain / Rcc01698-like, C-terminal / Protein of unknown function DUF2460 / Conserved hypothetical protein 2217 (DUF2460) / Baseplate hub protein, N-terminal attachment domain / Bacteriophage phiJL001, Gp84 / Bacteriophage phiJL001, Gp84, C-terminal / Phage conserved hypothetical protein BR0599 / Tip attachment protein J / Putative phage tail protein / Glycoside hydrolase superfamily Similarity search - Domain/homology | |||||||||
| Biological species |  Dinoroseobacter phage vB_DshS-R4C (virus) Dinoroseobacter phage vB_DshS-R4C (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.5 Å | |||||||||
 Authors Authors | Huang Y / Sun H / Wei S / Zheng Q / Li S / Zhang R / Xia N | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structure and proposed DNA delivery mechanism of a marine roseophage. Authors: Yang Huang / Hui Sun / Shuzhen Wei / Lanlan Cai / Liqin Liu / Yanan Jiang / Jiabao Xin / Zhenqin Chen / Yuqiong Que / Zhibo Kong / Tingting Li / Hai Yu / Jun Zhang / Ying Gu / Qingbing Zheng ...Authors: Yang Huang / Hui Sun / Shuzhen Wei / Lanlan Cai / Liqin Liu / Yanan Jiang / Jiabao Xin / Zhenqin Chen / Yuqiong Que / Zhibo Kong / Tingting Li / Hai Yu / Jun Zhang / Ying Gu / Qingbing Zheng / Shaowei Li / Rui Zhang / Ningshao Xia /  Abstract: Tailed bacteriophages (order, Caudovirales) account for the majority of all phages. However, the long flexible tail of siphophages hinders comprehensive investigation of the mechanism of viral gene ...Tailed bacteriophages (order, Caudovirales) account for the majority of all phages. However, the long flexible tail of siphophages hinders comprehensive investigation of the mechanism of viral gene delivery. Here, we report the atomic capsid and in-situ structures of the tail machine of the marine siphophage, vB_DshS-R4C (R4C), which infects Roseobacter. The R4C virion, comprising 12 distinct structural protein components, has a unique five-fold vertex of the icosahedral capsid that allows genome delivery. The specific position and interaction pattern of the tail tube proteins determine the atypical long rigid tail of R4C, and further provide negative charge distribution within the tail tube. A ratchet mechanism assists in DNA transmission, which is initiated by an absorption device that structurally resembles the phage-like particle, RcGTA. Overall, these results provide in-depth knowledge into the intact structure and underlining DNA delivery mechanism for the ecologically important siphophages. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34249.map.gz emd_34249.map.gz | 229.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34249-v30.xml emd-34249-v30.xml emd-34249.xml emd-34249.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34249.png emd_34249.png | 39.1 KB | ||
| Filedesc metadata |  emd-34249.cif.gz emd-34249.cif.gz | 6.9 KB | ||
| Others |  emd_34249_half_map_1.map.gz emd_34249_half_map_1.map.gz emd_34249_half_map_2.map.gz emd_34249_half_map_2.map.gz | 226.2 MB 226.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34249 http://ftp.pdbj.org/pub/emdb/structures/EMD-34249 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34249 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34249 | HTTPS FTP |
-Validation report
| Summary document |  emd_34249_validation.pdf.gz emd_34249_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34249_full_validation.pdf.gz emd_34249_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_34249_validation.xml.gz emd_34249_validation.xml.gz | 16.4 KB | Display | |
| Data in CIF |  emd_34249_validation.cif.gz emd_34249_validation.cif.gz | 18.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34249 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34249 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34249 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34249 | HTTPS FTP |
-Related structure data
| Related structure data |  8gtcMC  8gtaC  8gtbC  8gtdC  8gtfC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34249.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34249.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.12 Å | ||||||||||||||||||||||||||||||||||||
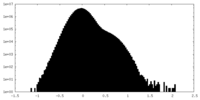
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34249_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
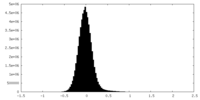
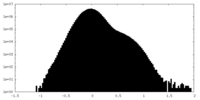
| Density Histograms |
-Half map: #1
| File | emd_34249_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
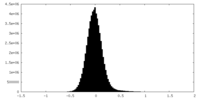
| Density Histograms |
- Sample components
Sample components
-Entire : Dinoroseobacter phage vB_DshS-R4C
| Entire | Name:  Dinoroseobacter phage vB_DshS-R4C (virus) Dinoroseobacter phage vB_DshS-R4C (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Dinoroseobacter phage vB_DshS-R4C
| Supramolecule | Name: Dinoroseobacter phage vB_DshS-R4C / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 2590919 / Sci species name: Dinoroseobacter phage vB_DshS-R4C / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Dinoroseobacter shibae DFL 12 = DSM 16493 (bacteria) Dinoroseobacter shibae DFL 12 = DSM 16493 (bacteria) |
-Macromolecule #1: Major tail protein
| Macromolecule | Name: Major tail protein / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Dinoroseobacter phage vB_DshS-R4C (virus) Dinoroseobacter phage vB_DshS-R4C (virus) |
| Molecular weight | Theoretical: 13.574816 KDa |
| Sequence | String: MLKGKDGVVK NASTGDSIGH LQSWALDTQR DEVSGWGMGD DAERAFTTVG RASGNFEVYL DPADPSDDLE PGDLVDLELY PGGESTGSG YRSVAGALIL STAESASKDG IPMLTVNWRT SGALPQKATV S UniProtKB: Major tail protein |
-Macromolecule #2: Distal tail protein
| Macromolecule | Name: Distal tail protein / type: protein_or_peptide / ID: 2 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Dinoroseobacter phage vB_DshS-R4C (virus) Dinoroseobacter phage vB_DshS-R4C (virus) |
| Molecular weight | Theoretical: 23.378309 KDa |
| Sequence | String: MQFIDVEFPR DIAAGCQAVL TRRDEVVTLA SGREEVNSRW ADTRRSWDAG LGVRDEADLA QVVALFEEVR GRLYAFRFRD WLDWRTAAT RAPITATDQP LGLGDGSRTA FQVVKVYGAV NPYTRPLSLP HPGTVRVALD GVTQPSGWTL TAPGGVITFD T PPALGVTV ...String: MQFIDVEFPR DIAAGCQAVL TRRDEVVTLA SGREEVNSRW ADTRRSWDAG LGVRDEADLA QVVALFEEVR GRLYAFRFRD WLDWRTAAT RAPITATDQP LGLGDGSRTA FQVVKVYGAV NPYTRPLSLP HPGTVRVALD GVTQPSGWTL TAPGGVITFD T PPALGVTV TAGCSFDVPV RFSDPELAVQ WAYFREGQAG LAQAPSIPLI EVRLDP UniProtKB: Tail protein |
-Macromolecule #3: Megatron protein
| Macromolecule | Name: Megatron protein / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Dinoroseobacter phage vB_DshS-R4C (virus) Dinoroseobacter phage vB_DshS-R4C (virus) |
| Molecular weight | Theoretical: 154.569125 KDa |
| Sequence | String: MATVLLAAAG GAIGTSLGGA LFGISAAVIG QAVGGLIGQT IDARLSGGGT IKQTGPRLDS LEVMVSQEGT PLADISGRVA VAGTVIWAT KLEEIARTTS TRVGSGKSSQ KVKSTNFDYA ASFAVSLGEG PLNGIGRIFL DGQVRDLSEM ISENRVRFYP G TEDQEPDP ...String: MATVLLAAAG GAIGTSLGGA LFGISAAVIG QAVGGLIGQT IDARLSGGGT IKQTGPRLDS LEVMVSQEGT PLADISGRVA VAGTVIWAT KLEEIARTTS TRVGSGKSSQ KVKSTNFDYA ASFAVSLGEG PLNGIGRIFL DGQVRDLSEM ISENRVRFYP G TEDQEPDP LIEAIEGAAP AFRGTAYLVF ERLFLSDFGN RVPQVRVEVF GQSGEMEKGV TGVCVIPGST EFGYMPAPVR QQ SLDGADV VWEGPENCNR YSRISDWSLS MDHLKNTLPA VQTVSLVVAW FGTDLRAGLC RFEPRIELRA KRTSIEWIAA GLN RGSATE VSRDENDRPA FGSSPADVSV VRAIMDLKAR GFRVVLYPFV MMDVTADQAL PSPAGTGAQG AYPWRGRIMP DVTG GRVSD QIAALMGTAQ PDDFTAAVTR NKSGEISQVN LTCPGAPGDA GLRRFILHLA KLADIAGGVD AFLVGTEFRG LSQAW EPQT YADAYEFLNS SEGWFAQNAA RTHEPGSLLV SATARDPRLI SPPLSIDGAA ARWIEIEFDR TSAPVWEGNV YYMTED HGF AGAFRKVISD AEAPADGTRA RMLLDMHDLT LGGDDWRTST ITRLRFDFSS ESDAVFRIHA IRLGGGRRYP FVDALRD LA QDVDTILPTA KISYAADWSE YHSHQDAGDL TFHLDPLWAD PAIDFVGIDN YLPLSDWRQG ADHLDYDATS GRTTPYDL D YLKSNIEGGE YWDWFYASQE DRDTQTRTPI SDGSYGEPWV FRQKAIRDWH ANAHHNRTAG VRATTSTGWV PGSKPVWFT EYGCPAVDLG ANRPNVFAAA SSSESALPWY SSGLRDDFMQ RQYLRAMAEW WTANGAPAVD LANCQAWAWD ARPFPEFPLR PATWSDGPD WRLGHWLNGR AGAAPAAEAI TRRAITRHGL ISADIDTSRA YGQADGYAAP APLGLGDYAQ PFEVALGLQT T ETGGALVI EAKPAAPLAA DVIEADLVDT PAIYTLTRGA LEDTPAAAIV RFRDGLSDYE ITAARARIGA GKEGGSATAD LA LVLDGDR GNAAAEMVLR AALTSRESLS VTLPRSATTL RPGSLVEVTL GTEARRLFLV DRVVDGQARE VTLRGFDRAA YAP SGGVFK AARAGRLQGS TSSLTRFLDL PLLPGVDAPE WEGFIAAHAE PWPGAMLHSR GSTPEGSFTL AAEADARATI GRTT AALPP APAHVWTPGP LVVTLFSGAL VGRPDLDVLD GANALAIQHP DGWEVVQFRE ATLTADRTWR LEGLLRGQRG TDGIV GPAP LPAGAAVVAI DTALVAAGLS ADDPGRALWW RSGPDAASLA AAPLRPHTFT AAGLRAFPPA HLRAVVTGGG DTSLSW VPR SRLVGTTWPD NGAPIPSGEG LERYQVTIGP TAAPVRVILA DTPAATYTAA ERAADGIAAP FRVAVSQISE TTGPGPW IE TIVTE UniProtKB: Tail protein |
-Macromolecule #4: Hub protein
| Macromolecule | Name: Hub protein / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Dinoroseobacter phage vB_DshS-R4C (virus) Dinoroseobacter phage vB_DshS-R4C (virus) |
| Molecular weight | Theoretical: 31.055357 KDa |
| Sequence | String: MTDYTDHLAT GCTTLARCYI LTRRDGVVMG FTDHDRDIDL DGTRCAAGAA LDASTAARSL GITPDDMDAG GALSADAITE ADLRAGRYD GAQVEVWEVN WTDPAVRGRL GVYTIGQVER GPLAFRAELR TRPALWNRPE GRIHTALCDV DRLGDHRCKL A LGPWQSAA ...String: MTDYTDHLAT GCTTLARCYI LTRRDGVVMG FTDHDRDIDL DGTRCAAGAA LDASTAARSL GITPDDMDAG GALSADAITE ADLRAGRYD GAQVEVWEVN WTDPAVRGRL GVYTIGQVER GPLAFRAELR TRPALWNRPE GRIHTALCDV DRLGDHRCKL A LGPWQSAA TVIEADGADL IVSGLDETAS NIFDRGVLDW TGGANAGTGS DIRVARPVAG GVRVSLWSAP PFPITAGDTA NA TVGCDRT ADTCRNRFDN LANFRGFPLM PGESFISEYA RPGDPDQSGG SRYD UniProtKB: Gene transfer agent |
-Macromolecule #5: Ribonuclease III
| Macromolecule | Name: Ribonuclease III / type: protein_or_peptide / ID: 5 / Number of copies: 9 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Dinoroseobacter phage vB_DshS-R4C (virus) Dinoroseobacter phage vB_DshS-R4C (virus) |
| Molecular weight | Theoretical: 22.783799 KDa |
| Sequence | String: MPTPFLALPL IAEGQAGQHV THNEALDMID ALAPRVVLSD SLATPPPAPP DRSAWIVPPG GSGFGGAGPG QIALRLGGVW HAITPAQGA RWRVLDRGAA VIWSGTSWRP ADVSGALGST LGLATIEATV TATGPSVTAP ALIPPRAIVL GVTSWTVQAV T GATSYRVG ...String: MPTPFLALPL IAEGQAGQHV THNEALDMID ALAPRVVLSD SLATPPPAPP DRSAWIVPPG GSGFGGAGPG QIALRLGGVW HAITPAQGA RWRVLDRGAA VIWSGTSWRP ADVSGALGST LGLATIEATV TATGPSVTAP ALIPPRAIVL GVTSWTVQAV T GATSYRVG VPGEPDKFGA SLGAAPGSSN IGVVGPFATY APTDVVVTAE GADFTGGTIG LAASVILPGA PV UniProtKB: Ribonuclease III |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER / Details: trRosetta server |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 4.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 11598 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)