[English] 日本語
 Yorodumi
Yorodumi- EMDB-33865: The Cryo-EM Structure of Human Tissue Nonspecific Alkaline Phosph... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The Cryo-EM Structure of Human Tissue Nonspecific Alkaline Phosphatase and Single-Chain Fragment Variable (ScFv) Complex. | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | alkaline phosphatase / bone mineralization / catalytic network / hypophosphatasia / structural biology / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationphosphoamidase / phosphoamidase activity / phosphoethanolamine phosphatase activity / pyridoxal phosphate metabolic process / response to vitamin B6 / futile creatine cycle / pyridoxal phosphatase activity / developmental process involved in reproduction / ADP phosphatase activity / response to macrophage colony-stimulating factor ...phosphoamidase / phosphoamidase activity / phosphoethanolamine phosphatase activity / pyridoxal phosphate metabolic process / response to vitamin B6 / futile creatine cycle / pyridoxal phosphatase activity / developmental process involved in reproduction / ADP phosphatase activity / response to macrophage colony-stimulating factor / inhibition of non-skeletal tissue mineralization / pyrophosphatase activity / Post-translational modification: synthesis of GPI-anchored proteins / cementum mineralization / alkaline phosphatase / alkaline phosphatase activity / response to sodium phosphate / phosphate ion homeostasis / inorganic diphosphate phosphatase activity / endochondral ossification / response to vitamin D / cellular homeostasis / bone mineralization / cellular response to organic cyclic compound / calcium ion homeostasis / response to glucocorticoid / side of membrane / extracellular matrix / skeletal system development / mitochondrial membrane / response to insulin / mitochondrial intermembrane space / osteoblast differentiation / positive regulation of cold-induced thermogenesis / response to lipopolysaccharide / response to antibiotic / calcium ion binding / ATP hydrolysis activity / extracellular exosome / extracellular region / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
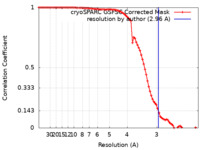
| Method | single particle reconstruction / cryo EM / Resolution: 2.96 Å | |||||||||
 Authors Authors | Yu YT / Yao DQ / Zhang Q / Rao B / Xia Y / Lu Y / Qin A / Ma PX / Cao Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: The structural pathology for hypophosphatasia caused by malfunctional tissue non-specific alkaline phosphatase. Authors: Yating Yu / Kewei Rong / Deqiang Yao / Qing Zhang / Xiankun Cao / Bing Rao / Ying Xia / Yi Lu / Yafeng Shen / Ying Yao / Hongtao Xu / Peixiang Ma / Yu Cao / An Qin /  Abstract: Hypophosphatasia (HPP) is a metabolic bone disease that manifests as developmental abnormalities in bone and dental tissues. HPP patients exhibit hypo-mineralization and osteopenia due to the ...Hypophosphatasia (HPP) is a metabolic bone disease that manifests as developmental abnormalities in bone and dental tissues. HPP patients exhibit hypo-mineralization and osteopenia due to the deficiency or malfunction of tissue non-specific alkaline phosphatase (TNAP), which catalyzes the hydrolysis of phosphate-containing molecules outside the cells, promoting the deposition of hydroxyapatite in the extracellular matrix. Despite the identification of hundreds of pathogenic TNAP mutations, the detailed molecular pathology of HPP remains unclear. Here, to address this issue, we determine the crystal structures of human TNAP at near-atomic resolution and map the major pathogenic mutations onto the structure. Our study reveals an unexpected octameric architecture for TNAP, which is generated by the tetramerization of dimeric TNAPs, potentially stabilizing the TNAPs in the extracellular environments. Moreover, we use cryo-electron microscopy to demonstrate that the TNAP agonist antibody (JTALP001) forms a stable complex with TNAP by binding to the octameric interface. The administration of JTALP001 enhances osteoblast mineralization and promoted recombinant TNAP-rescued mineralization in TNAP knockout osteoblasts. Our findings elucidate the structural pathology of HPP and highlight the therapeutic potential of the TNAP agonist antibody for osteoblast-associated bone disorders. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33865.map.gz emd_33865.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33865-v30.xml emd-33865-v30.xml emd-33865.xml emd-33865.xml | 16.7 KB 16.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33865_fsc.xml emd_33865_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_33865.png emd_33865.png | 89.2 KB | ||
| Others |  emd_33865_half_map_1.map.gz emd_33865_half_map_1.map.gz emd_33865_half_map_2.map.gz emd_33865_half_map_2.map.gz | 59.2 MB 59.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33865 http://ftp.pdbj.org/pub/emdb/structures/EMD-33865 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33865 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33865 | HTTPS FTP |
-Validation report
| Summary document |  emd_33865_validation.pdf.gz emd_33865_validation.pdf.gz | 914.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33865_full_validation.pdf.gz emd_33865_full_validation.pdf.gz | 914.5 KB | Display | |
| Data in XML |  emd_33865_validation.xml.gz emd_33865_validation.xml.gz | 16.5 KB | Display | |
| Data in CIF |  emd_33865_validation.cif.gz emd_33865_validation.cif.gz | 21.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33865 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33865 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33865 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33865 | HTTPS FTP |
-Related structure data
| Related structure data |  7yixMC  7yivC  7yiwC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33865.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33865.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_33865_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
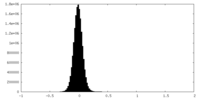
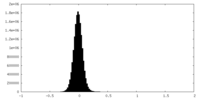
| Density Histograms |
-Half map: #1
| File | emd_33865_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
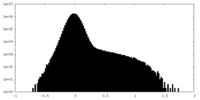
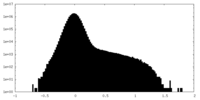
| Density Histograms |
- Sample components
Sample components
-Entire : hALPL-scFv
| Entire | Name: hALPL-scFv |
|---|---|
| Components |
|
-Supramolecule #1: hALPL-scFv
| Supramolecule | Name: hALPL-scFv / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Alkaline phosphatase, tissue-nonspecific isozyme
| Macromolecule | Name: Alkaline phosphatase, tissue-nonspecific isozyme / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: alkaline phosphatase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 57.020188 KDa |
| Recombinant expression | Organism:  Trichopalpus nigribasis (fry) Trichopalpus nigribasis (fry) |
| Sequence | String: MKTIIALSYI FCLVFAGRAL VPEKEKDPKY WRDQAQETLK YALELQKLNT NVAKNVIMFL GDGMGVSTVT AARILKGQLH HNPGEETRL EMDKFPFVAL SKTYNTNAQV PDSAGTATAY LCGVKANEGT VGVSAATERS RCNTTQGNEV TSILRWAKDA G KSVGIVTT ...String: MKTIIALSYI FCLVFAGRAL VPEKEKDPKY WRDQAQETLK YALELQKLNT NVAKNVIMFL GDGMGVSTVT AARILKGQLH HNPGEETRL EMDKFPFVAL SKTYNTNAQV PDSAGTATAY LCGVKANEGT VGVSAATERS RCNTTQGNEV TSILRWAKDA G KSVGIVTT TRVNHATPSA AYAHSADRDW YSDNEMPPEA LSQGCKDIAY QLMHNIRDID VIMGGGRKYM YPKNKTDVEY ES DEKARGT RLDGLDLVDT WKSFKPRYKH SHFIWNRTEL LTLDPHNVDY LLGLFEPGDM QYELNRNNVT DPSLSEMVVV AIQ ILRKNP KGFFLLVEGG RIDHGHHEGK AKQALHEAVE MDRAIGQAGS LTSSEDTLTV VTADHSHVFT FGGYTPRGNS IFGL APMLS DTDKKPFTAI LYGNGPGYKV VGGERENVSM VDYAHNNYQA QSAVPLRHET HGGEDVAVFS KGPMAHLLHG VHEQN YVPH VMAYAACIGA NLGHCAPAAA AENLYFQGCC PGCC UniProtKB: Alkaline phosphatase, tissue-nonspecific isozyme |
-Macromolecule #2: scFv
| Macromolecule | Name: scFv / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.830195 KDa |
| Recombinant expression | Organism: Mammalia (mammals) |
| Sequence | String: MYRMQLLSCI ALSLALVTNS AAQPAMAQVQ LVQSGAEVKK PGASVKVSCK ASGYTFTSYG ISWVRQAPGQ GLEWMGWISA YNGNTNYAQ KLQGRVTMTT DTSTSTAYME LRSLRSDDTA VYYCARDKGW NSEGSLEYWG QGTLVTVSSG GGGSGGGGSG G GGSDIVMT ...String: MYRMQLLSCI ALSLALVTNS AAQPAMAQVQ LVQSGAEVKK PGASVKVSCK ASGYTFTSYG ISWVRQAPGQ GLEWMGWISA YNGNTNYAQ KLQGRVTMTT DTSTSTAYME LRSLRSDDTA VYYCARDKGW NSEGSLEYWG QGTLVTVSSG GGGSGGGGSG G GGSDIVMT QSPSSVSASV GDRVTITCRA SQGISNWLGW YQQKPGKAPK LLIYGASSLQ SGVPSRFSGS GSGTDFTLTI SS LQPEDFA TYFCQQAYSL PLTFGGGTKL EIKRGAAA |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 4 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #6: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 6 / Number of copies: 4 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #7: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 7 / Number of copies: 2 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)