[English] 日本語
 Yorodumi
Yorodumi- EMDB-33795: Structure of GluN1a E698C-GluN2D NMDA receptor in cystines crossl... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of GluN1a E698C-GluN2D NMDA receptor in cystines crosslinked state. | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | ion channel / cryo-EM structure / glutamate receptor / synaptic protein / ELECTRON TRANSPORT | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationexcitatory chemical synaptic transmission / regulation of sensory perception of pain / Synaptic adhesion-like molecules / cellular response to L-glutamate / propylene metabolic process / response to glycine / regulation of monoatomic cation transmembrane transport / voltage-gated monoatomic cation channel activity / Assembly and cell surface presentation of NMDA receptors / NMDA glutamate receptor activity ...excitatory chemical synaptic transmission / regulation of sensory perception of pain / Synaptic adhesion-like molecules / cellular response to L-glutamate / propylene metabolic process / response to glycine / regulation of monoatomic cation transmembrane transport / voltage-gated monoatomic cation channel activity / Assembly and cell surface presentation of NMDA receptors / NMDA glutamate receptor activity / Neurexins and neuroligins / NMDA selective glutamate receptor complex / calcium ion transmembrane import into cytosol / glutamate binding / protein heterotetramerization / positive regulation of calcium ion transport into cytosol / positive regulation of reactive oxygen species biosynthetic process / glycine binding / startle response / Negative regulation of NMDA receptor-mediated neuronal transmission / Unblocking of NMDA receptors, glutamate binding and activation / monoatomic cation transmembrane transport / regulation of neuronal synaptic plasticity / monoatomic cation transport / Long-term potentiation / excitatory synapse / ligand-gated monoatomic ion channel activity / positive regulation of excitatory postsynaptic potential / calcium ion homeostasis / glutamate-gated receptor activity / synaptic cleft / glutamate-gated calcium ion channel activity / presynaptic active zone membrane / ligand-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / EPHB-mediated forward signaling / ionotropic glutamate receptor signaling pathway / Ras activation upon Ca2+ influx through NMDA receptor / hippocampal mossy fiber to CA3 synapse / positive regulation of synaptic transmission, glutamatergic / regulation of membrane potential / adult locomotory behavior / excitatory postsynaptic potential / synaptic transmission, glutamatergic / synaptic membrane / long-term synaptic potentiation / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / postsynaptic density membrane / brain development / visual learning / regulation of synaptic plasticity / terminal bouton / synaptic vesicle / signaling receptor activity / amyloid-beta binding / RAF/MAP kinase cascade / chemical synaptic transmission / postsynaptic membrane / response to ethanol / dendritic spine / postsynaptic density / calmodulin binding / neuron projection / glutamatergic synapse / dendrite / calcium ion binding / synapse / endoplasmic reticulum membrane / protein-containing complex binding / cell surface / positive regulation of transcription by RNA polymerase II / plasma membrane / cytoplasm Similarity search - Function | ||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
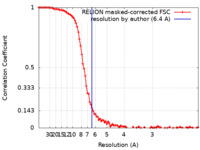
| Method | single particle reconstruction / cryo EM / Resolution: 6.4 Å | ||||||||||||||||||
 Authors Authors | Zhang JL / Zhu SJ / Zhang M | ||||||||||||||||||
| Funding support |  China, 5 items China, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits. Authors: Jilin Zhang / Ming Zhang / Qinrui Wang / Han Wen / Zheyi Liu / Fangjun Wang / Yuhang Wang / Fenyong Yao / Nan Song / Zengwei Kou / Yang Li / Fei Guo / Shujia Zhu /  Abstract: N-methyl-D-aspartate (NMDA) receptors are heterotetramers comprising two GluN1 and two alternate GluN2 (N2A-N2D) subunits. Here we report full-length cryo-EM structures of the human N1-N2D di- ...N-methyl-D-aspartate (NMDA) receptors are heterotetramers comprising two GluN1 and two alternate GluN2 (N2A-N2D) subunits. Here we report full-length cryo-EM structures of the human N1-N2D di-heterotetramer (di-receptor), rat N1-N2C di-receptor and N1-N2A-N2C tri-heterotetramer (tri-receptor) at a best resolution of 3.0 Å. The bilobate N-terminal domain (NTD) in N2D intrinsically adopts a closed conformation, leading to a compact NTD tetramer in the N1-N2D receptor. Additionally, crosslinking the ligand-binding domain (LBD) of two N1 protomers significantly elevated the channel open probability (Po) in N1-N2D di-receptors. Surprisingly, the N1-N2C di-receptor adopted both symmetric (minor) and asymmetric (major) conformations, the latter further locked by an allosteric potentiator, PYD-106, binding to a pocket between the NTD and LBD in only one N2C protomer. Finally, the N2A and N2C subunits in the N1-N2A-N2C tri-receptor display a conformation close to one protomer in the N1-N2A and N1-N2C di-receptors, respectively. These findings provide a comprehensive structural understanding of diverse function in major NMDA receptor subtypes. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33795.map.gz emd_33795.map.gz | 5.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33795-v30.xml emd-33795-v30.xml emd-33795.xml emd-33795.xml | 20.8 KB 20.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33795_fsc.xml emd_33795_fsc.xml | 10.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_33795.png emd_33795.png | 82.2 KB | ||
| Others |  emd_33795_half_map_1.map.gz emd_33795_half_map_1.map.gz emd_33795_half_map_2.map.gz emd_33795_half_map_2.map.gz | 64.1 MB 64.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33795 http://ftp.pdbj.org/pub/emdb/structures/EMD-33795 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33795 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33795 | HTTPS FTP |
-Validation report
| Summary document |  emd_33795_validation.pdf.gz emd_33795_validation.pdf.gz | 641.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33795_full_validation.pdf.gz emd_33795_full_validation.pdf.gz | 641.5 KB | Display | |
| Data in XML |  emd_33795_validation.xml.gz emd_33795_validation.xml.gz | 16.3 KB | Display | |
| Data in CIF |  emd_33795_validation.cif.gz emd_33795_validation.cif.gz | 20.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33795 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33795 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33795 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33795 | HTTPS FTP |
-Related structure data
| Related structure data |  7yfoMC  7yffC  7yfgC  7yfhC  7yfiC  7yflC  7yfmC  7yfrC  8hdkC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33795.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33795.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.071 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_33795_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_33795_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : NMDA receptor with NMDA 1 incorperated with NMDA 2D
| Entire | Name: NMDA receptor with NMDA 1 incorperated with NMDA 2D |
|---|---|
| Components |
|
-Supramolecule #1: NMDA receptor with NMDA 1 incorperated with NMDA 2D
| Supramolecule | Name: NMDA receptor with NMDA 1 incorperated with NMDA 2D / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Strain: Homo sapiens / Organ: brain / Tissue: brain / Organelle: synapse / Location in cell: plasma membrane Homo sapiens (human) / Strain: Homo sapiens / Organ: brain / Tissue: brain / Organelle: synapse / Location in cell: plasma membrane |
| Molecular weight | Theoretical: 384.54 kDa/nm |
-Macromolecule #1: Glutamate receptor ionotropic, NMDA 1
| Macromolecule | Name: Glutamate receptor ionotropic, NMDA 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 95.210102 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSTMRLLTLA LLFSCSVARA ACDPKIVNIG AVLSTRKHEQ MFREAVNQAN KRHGSWKIQL NATSVTHKPN AIQMALSVCE DLISSQVYA ILVSHPPTPN DHFTPTPVSY TAGFYRIPVL GLTTRMSIYS DKSIHLSFLR TVPPYSHQSS VWFEMMRVYS W NHIILLVS ...String: MSTMRLLTLA LLFSCSVARA ACDPKIVNIG AVLSTRKHEQ MFREAVNQAN KRHGSWKIQL NATSVTHKPN AIQMALSVCE DLISSQVYA ILVSHPPTPN DHFTPTPVSY TAGFYRIPVL GLTTRMSIYS DKSIHLSFLR TVPPYSHQSS VWFEMMRVYS W NHIILLVS DDHEGRAAQK RLETLLEERE SKAEKVLQFD PGTKNVTALL MEAKELEARV IILSASEDDA ATVYRAAAML NM TGSGYVW LVGEREISGN ALRYAPDGIL GLQLINGKNE SAHISDAVGV VAQAVHELLE KENITDPPRG CVGNTNIWKT GPL FKRVLM SSKYADGVTG RVEFNEDGDR KFANYSIMNL QNRKLVQVGI YNGTHVIPND RKIIWPGGET EKPRGYQMST RLKI VTIHQ EPFVYVKPTL SDGTCKEEFT VNGDPVKKVI CTGPNDTSPG SPRHTVPQCC YGFCIDLLIK LARTMNFTYE VHLVA DGKF GTQERVNNSN KKEWNGMMGE LLSGQADMIV APLTINNERA QYIEFSKPFK YQGLTILVKK EIPRSTLDSF MQPFQS TLW LLVGLSVHVV AVMLYLLDRF SPFGRFKVNS EEEEEDALTL SSAMWFSWGV LLNSGIGEGA PRSFSARILG MVWAGFA MI IVASYTANLA AFLVLDRPEE RITGINDPRL RNPSDKFIYA TVKQSSVDIY FRRQVCLSTM YRHMEKHNYE SAAEAIQA V RDNKLHAFIW DSAVLEFEAS QKCDLVTTGE LFFRSGFGIG MRKDSPWKQN VSLSILKSHE NGFMEDLDKT WVRYQECDS RSNAPATLTF ENMAGVFMLV AGGIVAGIFL IFIEIAYKRH KDARRKQ UniProtKB: Glutamate receptor ionotropic, NMDA 1 |
-Macromolecule #2: Glutamate receptor ionotropic, NMDA 2D
| Macromolecule | Name: Glutamate receptor ionotropic, NMDA 2D / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 97.226414 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MRGAGGPRGP RGPAKMLLLL ALACASPFPE EAPGPGGAGG PGGGLGGARP LNVALVFSGP AYAAEAARLG PAVAAAVRSP GLDVRPVAL VLNGSDPRSL VLQLCDLLSG LRVHGVVFED DSRAPAVAPI LDFLSAQTSL PIVAVHGGAA LVLTPKEKGS T FLQLGSST ...String: MRGAGGPRGP RGPAKMLLLL ALACASPFPE EAPGPGGAGG PGGGLGGARP LNVALVFSGP AYAAEAARLG PAVAAAVRSP GLDVRPVAL VLNGSDPRSL VLQLCDLLSG LRVHGVVFED DSRAPAVAPI LDFLSAQTSL PIVAVHGGAA LVLTPKEKGS T FLQLGSST EQQLQVIFEV LEEYDWTSFV AVTTRAPGHR AFLSYIEVLT DGSLVGWEHR GALTLDPGAG EAVLSAQLRS VS AQIRLLF CAREEAEPVF RAAEEAGLTG SGYVWFMVGP QLAGGGGSGA PGEPPLLPGG APLPAGLFAV RSAGWRDDLA RRV AAGVAV VARGAQALLR DYGFLPELGH DCRAQNRTHR GESLHRYFMN ITWDNRDYSF NEDGFLVNPS LVVISLTRDR TWEV VGSWE QQTLRLKYPL WSRYGRFLQP VDDTQHLTVA TLEERPFVIV EPADPISGTC IRDSVPCRSQ LNRTHSPPPD APRPE KRCC KGFCIDILKR LAHTIGFSYD LYLVTNGKHG KKIDGVWNGM IGEVFYQRAD MAIGSLTINE ERSEIVDFSV PFVETG ISV MVARSNGTVS PSAFLEPYSP AVWVMMFVMC LTVVAVTVFI FEYLSPVGYN RSLATGKRPG GSTFTIGKSI WLLWALV FN NSVPVENPRG TTSKIMVLVW AFFAVIFLAS YTANLAAFMI QEEYVDTVSG LSDRKFQRPQ EQYPPLKFGT VPNGSTEK N IRSNYPDMHS YMVRYNQPRV EEALTQLKAG KLDAFIYDAA VLNYMARKDE GCKLVTIGSG KVFATTGYGI ALHKGSRWK RPIDLALLQF LGDDEIEMLE RLWLSGICHN DKIEVMSSKL DIDNMAGVFY MLLVAMGLSL LVFAWEHLVY WRLRHCLGPA ASAWSHPQF EK UniProtKB: Glutamate receptor ionotropic, NMDA 2D |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.0 mg/mL |
|---|---|
| Buffer | pH: 8 / Component - Concentration: 150.0 mM / Component - Formula: NaCl / Component - Name: sodium chloride |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 101.325 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV / Details: Blot for 3 seconds before plunging. |
| Details | This sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: DIRECT ELECTRON DE-10 (5k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 5088 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 220 / Target criteria: Correlation coefficient |
| Output model |  PDB-7yfo: |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)