[English] 日本語
 Yorodumi
Yorodumi- EMDB-3330: Cryo-electron microscopy structure of AcrBA-TolC-MacA hybrid complex -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3330 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-electron microscopy structure of AcrBA-TolC-MacA hybrid complex | |||||||||
 Map data Map data | Cryo-electron microscopy single particle analysis 3D reconstruction of AcrBA-TolC-MacA hybrid complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Multidrug efflux pump / AcrA / AcrB / TolC / MacA / Resistance-nodulation-division transporter | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 8.2 Å | |||||||||
 Authors Authors | Jeong H / Kim J-S / Song S / Shigematsu H / Yokoyama T / Hyun J / Ha N-C | |||||||||
 Citation Citation |  Journal: Structure / Year: 2016 Journal: Structure / Year: 2016Title: Pseudoatomic Structure of the Tripartite Multidrug Efflux Pump AcrAB-TolC Reveals the Intermeshing Cogwheel-like Interaction between AcrA and TolC. Authors: Hyeongseop Jeong / Jin-Sik Kim / Saemee Song / Hideki Shigematsu / Takeshi Yokoyama / Jaekyung Hyun / Nam-Chul Ha /   Abstract: The resistance-nodulation-division type tripartite pump AcrAB-TolC and its homologs are responsible for multidrug resistance in Gram-negative bacteria by expelling a wide variety of toxic substrates. ...The resistance-nodulation-division type tripartite pump AcrAB-TolC and its homologs are responsible for multidrug resistance in Gram-negative bacteria by expelling a wide variety of toxic substrates. The three essential components, AcrA, AcrB, and TolC, must function in concert with each respective binding partner within the complex. In this study, we report an 8.2-Å resolution cryo-electron microscopy (cryo-EM) 3D reconstruction of the complex that consists of an AcrAB fusion protein and a chimeric TolC protein. The pseudoatomic structure derived from the cryo-EM reconstruction clearly demonstrates a model only compatible with the adaptor bridging mechanism, wherein the funnel-like AcrA hexamer forms an intermeshing cogwheel-like interaction with the α-barrel tip region of TolC. These observations provide a structural milestone for understanding multidrug resistance in pathogenic Gram-negative bacteria, and may also lead to the design of new antibacterial drugs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3330.map.gz emd_3330.map.gz | 4.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3330-v30.xml emd-3330-v30.xml emd-3330.xml emd-3330.xml | 10.3 KB 10.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3330.tif emd_3330.tif | 919.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3330 http://ftp.pdbj.org/pub/emdb/structures/EMD-3330 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3330 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3330 | HTTPS FTP |
-Validation report
| Summary document |  emd_3330_validation.pdf.gz emd_3330_validation.pdf.gz | 200.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3330_full_validation.pdf.gz emd_3330_full_validation.pdf.gz | 199.7 KB | Display | |
| Data in XML |  emd_3330_validation.xml.gz emd_3330_validation.xml.gz | 6.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3330 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3330 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3330 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3330 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3330.map.gz / Format: CCP4 / Size: 51.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3330.map.gz / Format: CCP4 / Size: 51.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-electron microscopy single particle analysis 3D reconstruction of AcrBA-TolC-MacA hybrid complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.947 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
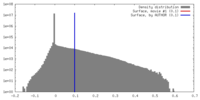
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : AcrBA fusion protein in complex with MacA-TolC hybrid dimer protein
| Entire | Name: AcrBA fusion protein in complex with MacA-TolC hybrid dimer protein |
|---|---|
| Components |
|
-Supramolecule #1000: AcrBA fusion protein in complex with MacA-TolC hybrid dimer protein
| Supramolecule | Name: AcrBA fusion protein in complex with MacA-TolC hybrid dimer protein type: sample / ID: 1000 Oligomeric state: AcrBA homotrimer in complex with MacA-TolC homotrimer Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 650 KDa |
-Macromolecule #1: AcrBA-TolC-MacA hybrid complex
| Macromolecule | Name: AcrBA-TolC-MacA hybrid complex / type: protein_or_peptide / ID: 1 Details: 3 copies of AcrBA fusion protein in complex with 3 copies of TolC-MacA hybrid dimer Number of copies: 6 / Oligomeric state: Hexamer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 650 KDa |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.15 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 20 mM HEPES, 150 mM NaCl, and 2 mM b-mercaptoethanol |
| Grid | Details: Holey carbon EM grid (Quantifoil R1.2/1.3) |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Instrument: FEI VITROBOT MARK IV Method: Blot force 2, Blot time 4 sec, Wait time 30 sec, No drain time, Chamber temperature 4 degrees Celcius |
- Electron microscopy
Electron microscopy
| Microscope | OTHER |
|---|---|
| Date | Apr 4, 2015 |
| Image recording | Category: CCD / Film or detector model: FEI FALCON II (4k x 4k) / Number real images: 595 / Average electron dose: 24 e/Å2 Details: Every image is the average of seven frames recorded by direct electron detector |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 71895 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 53000 |
| Sample stage | Specimen holder model: OTHER |
- Image processing
Image processing
| Details | Image processing was performed using Relion 1.3 software. Particles were semi-automatically selected. From 200 class averages, particles that belong in class averages with sharp structural details were used for subsequent 3D classification and refinement. After refinement, B-factor sharpening and MTF correction was performed. |
|---|---|
| CTF correction | Details: each particle |
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 8.2 Å / Resolution method: OTHER / Software - Name: Relion / Number images used: 20402 |
| Final two d classification | Number classes: 200 |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name: Chimera, PHENIX, Coot |
| Details | Experimental and model structures of AcrB timer, AcrA hexamer and MacA-TolC hybrid dimer were docked into cryo-EM reconstruction as separate rigid bodies. Docking was manually performed using UCSF Chimera and PHENIX, followed by manual adjustments using Coot. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)