+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of RuvA-RuvB-Holliday junction complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Holliday junction / Homologous recombination / DNA damage repair / Branch migration / RECOMBINATION-DNA COMPLEX | |||||||||
| Function / homology |  Function and homology information Function and homology informationcellular response to chromate / Holliday junction helicase complex / cellular response to selenite ion / Holliday junction resolvase complex / four-way junction helicase activity / four-way junction DNA binding / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / DNA recombination / DNA repair / ATP hydrolysis activity ...cellular response to chromate / Holliday junction helicase complex / cellular response to selenite ion / Holliday junction resolvase complex / four-way junction helicase activity / four-way junction DNA binding / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / DNA recombination / DNA repair / ATP hydrolysis activity / ATP binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Pseudomonas aeruginosa PAO1 (bacteria) / synthetic construct (others) Pseudomonas aeruginosa PAO1 (bacteria) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.02 Å | |||||||||
 Authors Authors | Lin Z / Qu Q / Zhang X / Zhou Z | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Front Plant Sci / Year: 2023 Journal: Front Plant Sci / Year: 2023Title: Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from . Authors: Xu Zhang / Zixuan Zhou / Lin Dai / Yulin Chao / Zheng Liu / Mingdong Huang / Qianhui Qu / Zhonghui Lin /  Abstract: Holliday junction (HJ) is a four-way structured DNA intermediate in homologous recombination. In bacteria, the HJ-specific binding protein RuvA and the motor protein RuvB together form the RuvAB ...Holliday junction (HJ) is a four-way structured DNA intermediate in homologous recombination. In bacteria, the HJ-specific binding protein RuvA and the motor protein RuvB together form the RuvAB complex to catalyze HJ branch migration. (, Pa) is a ubiquitous opportunistic bacterial pathogen that can cause serious infection in a variety of host species, including vertebrate animals, insects and plants. Here, we describe the cryo-Electron Microscopy (cryo-EM) structure of the RuvAB-HJ intermediate complex from . The structure shows that two RuvA tetramers sandwich HJ at the junction center and disrupt base pairs at the branch points of RuvB-free HJ arms. Eight RuvB subunits are recruited by the RuvA octameric core and form two open-rings to encircle two opposite HJ arms. Each RuvB subunit individually binds a RuvA domain III. The four RuvB subunits within the ring display distinct subdomain conformations, and two of them engage the central DNA duplex at both strands with their C-terminal β-hairpins. Together with the biochemical analyses, our structure implicates a potential mechanism of RuvB motor assembly onto HJ DNA. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33044.map.gz emd_33044.map.gz | 398.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33044-v30.xml emd-33044-v30.xml emd-33044.xml emd-33044.xml | 21.7 KB 21.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33044.png emd_33044.png | 33.2 KB | ||
| Others |  emd_33044_half_map_1.map.gz emd_33044_half_map_1.map.gz emd_33044_half_map_2.map.gz emd_33044_half_map_2.map.gz | 391.8 MB 391.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33044 http://ftp.pdbj.org/pub/emdb/structures/EMD-33044 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33044 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33044 | HTTPS FTP |
-Validation report
| Summary document |  emd_33044_validation.pdf.gz emd_33044_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33044_full_validation.pdf.gz emd_33044_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_33044_validation.xml.gz emd_33044_validation.xml.gz | 17.7 KB | Display | |
| Data in CIF |  emd_33044_validation.cif.gz emd_33044_validation.cif.gz | 20.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33044 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33044 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33044 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33044 | HTTPS FTP |
-Related structure data
| Related structure data |  7x7qMC  7x5aC  7x5bC  7x7pC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33044.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33044.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
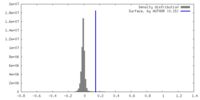
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_33044_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
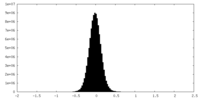
| Density Histograms |
-Half map: #1
| File | emd_33044_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
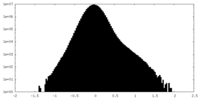
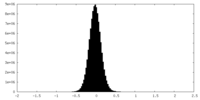
| Density Histograms |
- Sample components
Sample components
-Entire : RuvA-RuvB-Holliday junction complex
| Entire | Name: RuvA-RuvB-Holliday junction complex |
|---|---|
| Components |
|
-Supramolecule #1: RuvA-RuvB-Holliday junction complex
| Supramolecule | Name: RuvA-RuvB-Holliday junction complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria) |
-Supramolecule #2: RuvA-RuvB
| Supramolecule | Name: RuvA-RuvB / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Supramolecule #3: DNA
| Supramolecule | Name: DNA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3-#6 |
|---|
-Macromolecule #1: Holliday junction ATP-dependent DNA helicase RuvA
| Macromolecule | Name: Holliday junction ATP-dependent DNA helicase RuvA / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO / EC number: DNA helicase |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) / Strain: PAO1 Pseudomonas aeruginosa PAO1 (bacteria) / Strain: PAO1 |
| Molecular weight | Theoretical: 21.987594 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIGRLRGTLA EKQPPHLILD VNGVGYEVEV PMTTLYRLPS VGEPVTLHTH LVVREDAHLL YGFAEKRERE LFRELIRLNG VGPKLALAL MSGLEVDELV RCVQAQDTST LVKIPGVGKK TAERLLVELK DRFKAWENMP TIAPLVMEPR ASATVSSAEA D AVSALIAL ...String: MIGRLRGTLA EKQPPHLILD VNGVGYEVEV PMTTLYRLPS VGEPVTLHTH LVVREDAHLL YGFAEKRERE LFRELIRLNG VGPKLALAL MSGLEVDELV RCVQAQDTST LVKIPGVGKK TAERLLVELK DRFKAWENMP TIAPLVMEPR ASATVSSAEA D AVSALIAL GFKPQEASRA VAAVPGEDLS SEEMIRQALK GMV UniProtKB: Holliday junction branch migration complex subunit RuvA |
-Macromolecule #2: Holliday junction ATP-dependent DNA helicase RuvB
| Macromolecule | Name: Holliday junction ATP-dependent DNA helicase RuvB / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO / EC number: DNA helicase |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) / Strain: PAO1 Pseudomonas aeruginosa PAO1 (bacteria) / Strain: PAO1 |
| Molecular weight | Theoretical: 38.981578 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIEPDRLISA VSGRERDEQL DRAIRPLKLA DYIGQPSVRE QMELFIHAAR GRQEALDHTL IFGPPGLGKT TLANIIAQEM GVSIKSTSG PVLERPGDLA ALLTNLEAGD VLFVDEIHRL SPIVEEVLYP AMEDFQLDIM IGEGPAARSI KLDLPPFTLV G ATTRAGML ...String: MIEPDRLISA VSGRERDEQL DRAIRPLKLA DYIGQPSVRE QMELFIHAAR GRQEALDHTL IFGPPGLGKT TLANIIAQEM GVSIKSTSG PVLERPGDLA ALLTNLEAGD VLFVDEIHRL SPIVEEVLYP AMEDFQLDIM IGEGPAARSI KLDLPPFTLV G ATTRAGML TNPLRDRFGI VQRLEFYNVE DLATIVSRSA GILGLEIEPQ GAAEIAKRAR GTPRIANRLL RRVRDFAEVR GQ GDITRVI ADKALNLLDV DERGFDHLDR RLLLTMIDKF DGGPVGIDNL AAALSEERHT IEDVLEPYLI QQGYIMRTPR GRV VTRHAY LHFGLNIPKR LGPGVTTDLF TSEDGN UniProtKB: Holliday junction branch migration complex subunit RuvB |
-Macromolecule #3: DNA (40-MER)
| Macromolecule | Name: DNA (40-MER) / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 12.321062 KDa |
| Sequence | String: (DA)(DT)(DA)(DT)(DT)(DA)(DT)(DA)(DA)(DT) (DA)(DT)(DA)(DT)(DA)(DA)(DT)(DA)(DA)(DT) (DA)(DA)(DA)(DT)(DA)(DT)(DT)(DT)(DA) (DA)(DT)(DA)(DT)(DT)(DA)(DT)(DA)(DA)(DT) (DA) |
-Macromolecule #4: DNA (40-MER)
| Macromolecule | Name: DNA (40-MER) / type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 12.303033 KDa |
| Sequence | String: (DA)(DA)(DA)(DA)(DT)(DT)(DA)(DA)(DT)(DA) (DT)(DT)(DA)(DA)(DA)(DT)(DA)(DT)(DT)(DT) (DA)(DT)(DT)(DA)(DT)(DT)(DA)(DT)(DA) (DT)(DA)(DT)(DT)(DA)(DT)(DA)(DA)(DT)(DA) (DT) |
-Macromolecule #5: DNA (26-MER)
| Macromolecule | Name: DNA (26-MER) / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 7.972225 KDa |
| Sequence | String: (DT)(DA)(DT)(DT)(DA)(DT)(DA)(DA)(DT)(DA) (DT)(DT)(DA)(DA)(DA)(DT)(DA)(DT)(DT)(DA) (DT)(DA)(DT)(DT)(DT)(DA) |
-Macromolecule #6: DNA (26-MER)
| Macromolecule | Name: DNA (26-MER) / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 7.972224 KDa |
| Sequence | String: (DT)(DA)(DA)(DA)(DT)(DA)(DT)(DA)(DA)(DT) (DA)(DT)(DT)(DT)(DA)(DA)(DT)(DA)(DT)(DT) (DA)(DA)(DT)(DT)(DT)(DT) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 260.0 kPa |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated magnification: 60241 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: FOURIER SPACE / Resolution.type: BY AUTHOR / Resolution: 7.02 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 3.1) / Number images used: 20536 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. 3.1) |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-7x7q: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)