+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | DNA bound-ICP1 Csy complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR-Cas system / RNA BINDING PROTEIN / RNA BINDING PROTEIN-RNA-DNA complex | |||||||||
| Function / homology | CRISPR-associated protein Csy2 / CRISPR-associated protein (Cas_Csy2) / CRISPR-associated protein Csy3 / CRISPR-associated protein (Cas_Csy3) / Csy3 / Csy2 / Csy1 Function and homology information Function and homology information | |||||||||
| Biological species |  Vibrio phage ICP1_2011_A (virus) Vibrio phage ICP1_2011_A (virus) | |||||||||
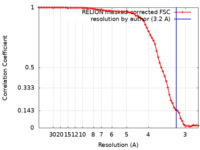
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Zhang M / Peng R | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Mechanistic insights into DNA binding and cleavage by a compact type I-F CRISPR-Cas system in bacteriophage. Authors: Manling Zhang / Ruchao Peng / Qi Peng / Sheng Liu / Zhiteng Li / Yuqin Zhang / Hao Song / Jia Yang / Xiao Xing / Peiyi Wang / Jianxun Qi / George F Gao /  Abstract: CRISPR-Cas systems are widespread adaptive antiviral systems used in prokaryotes. Some phages, in turn, although have small genomes can economize the use of genetic space to encode compact or ...CRISPR-Cas systems are widespread adaptive antiviral systems used in prokaryotes. Some phages, in turn, although have small genomes can economize the use of genetic space to encode compact or incomplete CRISPR-Cas systems to inhibit the host and establish infection. Phage ICP1, infecting , encodes a compact type I-F CRISPR-Cas system to suppress the antiphage mobile genetic element in the host genome. However, the mechanism by which this compact system recognizes the target DNA and executes interference remains elusive. Here, we present the electron cryo-microscopy (cryo-EM) structures of both apo- and DNA-bound ICP1 surveillance complexes (Aka Csy complex). Unlike most other type I surveillance complexes, the ICP1 Csy complex lacks the Cas11 subunit or a structurally homologous domain, which is crucial for dsDNA binding and Cas3 activation in other type I CRISPR-Cas systems. Structural and functional analyses revealed that the compact ICP1 Csy complex alone is inefficient in binding to dsDNA targets, presumably stalled at a partial R-loop conformation. The presence of Cas2/3 facilitates dsDNA binding and allows effective dsDNA target cleavage. Additionally, we found that Cas2/3 efficiently cleaved the dsDNA target presented by the ICP1 Csy complex, but not vice versa. These findings suggest a unique mechanism for target dsDNA binding and cleavage by the compact phage-derived CRISPR-Cas system. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32874.map.gz emd_32874.map.gz | 3.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32874-v30.xml emd-32874-v30.xml emd-32874.xml emd-32874.xml | 16.4 KB 16.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32874_fsc.xml emd_32874_fsc.xml | 7.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_32874.png emd_32874.png | 115.6 KB | ||
| Filedesc metadata |  emd-32874.cif.gz emd-32874.cif.gz | 6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32874 http://ftp.pdbj.org/pub/emdb/structures/EMD-32874 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32874 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32874 | HTTPS FTP |
-Validation report
| Summary document |  emd_32874_validation.pdf.gz emd_32874_validation.pdf.gz | 411 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32874_full_validation.pdf.gz emd_32874_full_validation.pdf.gz | 410.6 KB | Display | |
| Data in XML |  emd_32874_validation.xml.gz emd_32874_validation.xml.gz | 9.5 KB | Display | |
| Data in CIF |  emd_32874_validation.cif.gz emd_32874_validation.cif.gz | 12.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32874 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32874 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32874 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32874 | HTTPS FTP |
-Related structure data
| Related structure data |  7wwvMC  7wkoC  7wkpC  7wwuC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_32874.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32874.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.38 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : target DNA bound ICP1 Csy complex
| Entire | Name: target DNA bound ICP1 Csy complex |
|---|---|
| Components |
|
-Supramolecule #1: target DNA bound ICP1 Csy complex
| Supramolecule | Name: target DNA bound ICP1 Csy complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Vibrio phage ICP1_2011_A (virus) Vibrio phage ICP1_2011_A (virus) |
-Macromolecule #1: Csy1
| Macromolecule | Name: Csy1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vibrio phage ICP1_2011_A (virus) Vibrio phage ICP1_2011_A (virus) |
| Molecular weight | Theoretical: 22.84151 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGRENLYFQ GMIKEMIEDF ISKGGLIFTH SGRYTNTNNS CFIFNKNDIG VDTKVDMYTP KSAGIKNEEG ENLWQVLNK ANMFYRIYSG ELGEELQYLL KSCCTAKEDV TTLPQIYFKN GEGYDILVPI GNAHNLISGT EYLWEHKYYN T FTQKLGGS ...String: MGSSHHHHHH SSGRENLYFQ GMIKEMIEDF ISKGGLIFTH SGRYTNTNNS CFIFNKNDIG VDTKVDMYTP KSAGIKNEEG ENLWQVLNK ANMFYRIYSG ELGEELQYLL KSCCTAKEDV TTLPQIYFKN GEGYDILVPI GNAHNLISGT EYLWEHKYYN T FTQKLGGS NPQNCTHACN KMRGGFKQFN CTPPQVEDNY NA UniProtKB: Csy1 |
-Macromolecule #2: Csy2
| Macromolecule | Name: Csy2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vibrio phage ICP1_2011_A (virus) Vibrio phage ICP1_2011_A (virus) |
| Molecular weight | Theoretical: 29.833129 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGRENLYFQ GMRKFIIVKN VKVDGINAKS SDITVGMPPA TTFCGLGETM SIKTGIVVKA VSYGSVKFEV RGSRFNTSV TKFAWQDRGN GGKANNNSPI QPKPLADGVF TLCFEVEWED CAEVLVDKVT NFINTARIAG GTIASFNKPF V KVAKDAEE ...String: MGSSHHHHHH SSGRENLYFQ GMRKFIIVKN VKVDGINAKS SDITVGMPPA TTFCGLGETM SIKTGIVVKA VSYGSVKFEV RGSRFNTSV TKFAWQDRGN GGKANNNSPI QPKPLADGVF TLCFEVEWED CAEVLVDKVT NFINTARIAG GTIASFNKPF V KVAKDAEE LASVKNAMMP CYVVVDCGVE VNIFEDAVNR KLQPMVNGYK KLEKIVDNKH MRDKFTPAYL ATPTYTMIGY KM VSNVDNF DQALWQYGEN TKVKTIGGIY ND UniProtKB: Csy2 |
-Macromolecule #3: Csy3
| Macromolecule | Name: Csy3 / type: protein_or_peptide / ID: 3 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vibrio phage ICP1_2011_A (virus) Vibrio phage ICP1_2011_A (virus) |
| Molecular weight | Theoretical: 35.830867 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGRENLYFQ GMTKLKAPAV LAYSRKINPT NALMFAVNWS DRDNTTAVMV GTKTVAGTQS VRGNPNDADK GNIQTVNFA NLPHNKNTLL VKYNVKFVGD VFKAELGGGE YSNTLQTALE NTDFGTLAYR YVYNIAAGRT LWRNRVGAES I ETVITVND ...String: MGSSHHHHHH SSGRENLYFQ GMTKLKAPAV LAYSRKINPT NALMFAVNWS DRDNTTAVMV GTKTVAGTQS VRGNPNDADK GNIQTVNFA NLPHNKNTLL VKYNVKFVGD VFKAELGGGE YSNTLQTALE NTDFGTLAYR YVYNIAAGRT LWRNRVGAES I ETVITVND QTFTFSDLLV NEFDEDVDVA EIADMVAGVL SGEGFVTLKV EHYMLLGEGS EVFPSQEFVE NSKLSKQLFD LN GQAAMHD QKIGNAIRTI DTWYEDATTP IAVEPYGSVV RNGVAYRAGN KTDLFTLMDG AVNGKSLTEE DQMFVTANLI RGG VFGGGK D UniProtKB: Csy3 |
-Macromolecule #4: guide-RNA
| Macromolecule | Name: guide-RNA / type: rna / ID: 4 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Vibrio phage ICP1_2011_A (virus) Vibrio phage ICP1_2011_A (virus) |
| Molecular weight | Theoretical: 19.046227 KDa |
| Sequence | String: CUUAAAGAGU CAACCCUUUG CUUAUCUUCC CUAUUUAAAU GUUAGCAGCC GCAUAGGCUG GENBANK: GENBANK: KC152959.1 |
-Macromolecule #5: target strand DNA
| Macromolecule | Name: target strand DNA / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Vibrio phage ICP1_2011_A (virus) Vibrio phage ICP1_2011_A (virus) |
| Molecular weight | Theoretical: 18.571943 KDa |
| Sequence | String: (DC)(DG)(DT)(DT)(DT)(DA)(DC)(DA)(DG)(DC) (DA)(DA)(DT)(DT)(DT)(DA)(DA)(DA)(DT)(DA) (DG)(DG)(DG)(DA)(DA)(DG)(DA)(DT)(DA) (DA)(DG)(DC)(DA)(DA)(DA)(DG)(DG)(DG)(DT) (DT) (DG)(DA)(DC)(DG)(DA)(DA) ...String: (DC)(DG)(DT)(DT)(DT)(DA)(DC)(DA)(DG)(DC) (DA)(DA)(DT)(DT)(DT)(DA)(DA)(DA)(DT)(DA) (DG)(DG)(DG)(DA)(DA)(DG)(DA)(DT)(DA) (DA)(DG)(DC)(DA)(DA)(DA)(DG)(DG)(DG)(DT) (DT) (DG)(DA)(DC)(DG)(DA)(DA)(DA)(DG) (DC)(DC)(DC)(DT)(DT)(DT)(DG)(DT)(DC)(DC) (DC)(DT) |
-Macromolecule #6: non-target strand DNA
| Macromolecule | Name: non-target strand DNA / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Vibrio phage ICP1_2011_A (virus) Vibrio phage ICP1_2011_A (virus) |
| Molecular weight | Theoretical: 18.616916 KDa |
| Sequence | String: (DA)(DG)(DG)(DG)(DA)(DC)(DA)(DA)(DA)(DG) (DG)(DG)(DC)(DT)(DT)(DT)(DC)(DA)(DG)(DA) (DG)(DG)(DA)(DA)(DA)(DC)(DC)(DC)(DA) (DT)(DC)(DC)(DG)(DC)(DT)(DG)(DG)(DA)(DT) (DT) (DG)(DC)(DC)(DC)(DC)(DG) ...String: (DA)(DG)(DG)(DG)(DA)(DC)(DA)(DA)(DA)(DG) (DG)(DG)(DC)(DT)(DT)(DT)(DC)(DA)(DG)(DA) (DG)(DG)(DA)(DA)(DA)(DC)(DC)(DC)(DA) (DT)(DC)(DC)(DG)(DC)(DT)(DG)(DG)(DA)(DT) (DT) (DG)(DC)(DC)(DC)(DC)(DG)(DG)(DG) (DG)(DT)(DG)(DC)(DT)(DG)(DT)(DA)(DA)(DA) (DC)(DG) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)