+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7wkp | ||||||
|---|---|---|---|---|---|---|---|
| Title | ICP1 Csy4 | ||||||
 Components Components |
| ||||||
 Keywords Keywords | RNA BINDING PROTEIN/RNA / Complex / RNA binding protein / RNA BINDING PROTEIN-RNA complex | ||||||
| Function / homology | CRISPR-associated endoribonuclease Cas6/Csy4, subtype I-F/YPEST / CRISPR-associated endoribonuclease Cas6/Csy4, subtype I-F/YPEST superfamily / CRISPR-associated protein (Cas_Csy4) / maintenance of CRISPR repeat elements / endonuclease activity / : / RNA / RNA (> 10) / Csy4 Function and homology information Function and homology information | ||||||
| Biological species |  Vibrio phage ICP1_2011_A (virus) Vibrio phage ICP1_2011_A (virus) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2 Å MOLECULAR REPLACEMENT / Resolution: 2 Å | ||||||
 Authors Authors | Zhang, M. / Peng, R. | ||||||
| Funding support |  China, 1items China, 1items
| ||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Mechanistic insights into DNA binding and cleavage by a compact type I-F CRISPR-Cas system in bacteriophage. Authors: Manling Zhang / Ruchao Peng / Qi Peng / Sheng Liu / Zhiteng Li / Yuqin Zhang / Hao Song / Jia Yang / Xiao Xing / Peiyi Wang / Jianxun Qi / George F Gao /  Abstract: CRISPR-Cas systems are widespread adaptive antiviral systems used in prokaryotes. Some phages, in turn, although have small genomes can economize the use of genetic space to encode compact or ...CRISPR-Cas systems are widespread adaptive antiviral systems used in prokaryotes. Some phages, in turn, although have small genomes can economize the use of genetic space to encode compact or incomplete CRISPR-Cas systems to inhibit the host and establish infection. Phage ICP1, infecting , encodes a compact type I-F CRISPR-Cas system to suppress the antiphage mobile genetic element in the host genome. However, the mechanism by which this compact system recognizes the target DNA and executes interference remains elusive. Here, we present the electron cryo-microscopy (cryo-EM) structures of both apo- and DNA-bound ICP1 surveillance complexes (Aka Csy complex). Unlike most other type I surveillance complexes, the ICP1 Csy complex lacks the Cas11 subunit or a structurally homologous domain, which is crucial for dsDNA binding and Cas3 activation in other type I CRISPR-Cas systems. Structural and functional analyses revealed that the compact ICP1 Csy complex alone is inefficient in binding to dsDNA targets, presumably stalled at a partial R-loop conformation. The presence of Cas2/3 facilitates dsDNA binding and allows effective dsDNA target cleavage. Additionally, we found that Cas2/3 efficiently cleaved the dsDNA target presented by the ICP1 Csy complex, but not vice versa. These findings suggest a unique mechanism for target dsDNA binding and cleavage by the compact phage-derived CRISPR-Cas system. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7wkp.cif.gz 7wkp.cif.gz | 72.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7wkp.ent.gz pdb7wkp.ent.gz | 44 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7wkp.json.gz 7wkp.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/wk/7wkp https://data.pdbj.org/pub/pdb/validation_reports/wk/7wkp ftp://data.pdbj.org/pub/pdb/validation_reports/wk/7wkp ftp://data.pdbj.org/pub/pdb/validation_reports/wk/7wkp | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  7wkoC  7wwuC  7wwvC  2xliS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 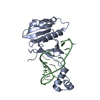
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 21168.271 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Vibrio phage ICP1_2011_A (virus) / Gene: csy4, ICP12011A_085 Vibrio phage ICP1_2011_A (virus) / Gene: csy4, ICP12011A_085Production host:  References: UniProt: M1R9H3 |
|---|---|
| #2: RNA chain | Mass: 19046.227 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Vibrio phage ICP1_2011_A (virus) Vibrio phage ICP1_2011_A (virus)Production host:  References: GenBank: 452118997 |
| #3: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.14 Å3/Da / Density % sol: 42.52 % |
|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, sitting drop Details: 0.4M Sodium thiocyanate, 0.1M Sodium acetate pH 4.0, 16% w/v PEG 4000 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRF SSRF  / Beamline: BL19U1 / Wavelength: 1.03923 Å / Beamline: BL19U1 / Wavelength: 1.03923 Å |
| Detector | Type: DECTRIS PILATUS3 6M / Detector: PIXEL / Date: Apr 1, 2019 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.03923 Å / Relative weight: 1 |
| Reflection | Resolution: 1.996→31.04 Å / Num. obs: 25046 / % possible obs: 98.4 % / Redundancy: 25.9 % / Biso Wilson estimate: 25.24 Å2 / Rmerge(I) obs: 0.145 / Net I/σ(I): 26.314 |
| Reflection shell | Resolution: 1.996→2.067 Å / Rmerge(I) obs: 1.88 / Num. unique obs: 2101 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 2XLI Resolution: 2→31.04 Å / SU ML: 0.2028 / Cross valid method: FREE R-VALUE / σ(F): 1.34 / Phase error: 23.2713 Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 29.6 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2→31.04 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller





 PDBj
PDBj






























