+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31420 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Marburg virus nucleoprotein-RNA complex | |||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | nucleoprotein / VIRAL PROTEIN | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationviral RNA genome packaging / helical viral capsid / viral budding via host ESCRT complex / viral nucleocapsid / host cell cytoplasm / ribonucleoprotein complex / RNA binding Similarity search - Function | |||||||||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Lake Victoria marburgvirus (strain Angola/2005) / synthetic construct (others) Lake Victoria marburgvirus (strain Angola/2005) / synthetic construct (others) | |||||||||||||||||||||
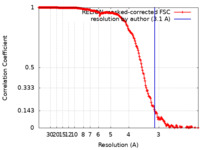
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||||||||||||||
 Authors Authors | Fujita FY / Sugita Y | |||||||||||||||||||||
| Funding support |  Japan, 6 items Japan, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural insight into Marburg virus nucleoprotein-RNA complex formation. Authors: Yoko Fujita-Fujiharu / Yukihiko Sugita / Yuki Takamatsu / Kazuya Houri / Manabu Igarashi / Yukiko Muramoto / Masahiro Nakano / Yugo Tsunoda / Ichiro Taniguchi / Stephan Becker / Takeshi Noda /   Abstract: The nucleoprotein (NP) of Marburg virus (MARV), a close relative of Ebola virus (EBOV), encapsidates the single-stranded, negative-sense viral genomic RNA (vRNA) to form the helical NP-RNA complex. ...The nucleoprotein (NP) of Marburg virus (MARV), a close relative of Ebola virus (EBOV), encapsidates the single-stranded, negative-sense viral genomic RNA (vRNA) to form the helical NP-RNA complex. The NP-RNA complex constitutes the core structure for the assembly of the nucleocapsid that is responsible for viral RNA synthesis. Although appropriate interactions among NPs and RNA are required for the formation of nucleocapsid, the structural basis of the helical assembly remains largely elusive. Here, we show the structure of the MARV NP-RNA complex determined using cryo-electron microscopy at a resolution of 3.1 Å. The structures of the asymmetric unit, a complex of an NP and six RNA nucleotides, was very similar to that of EBOV, suggesting that both viruses share common mechanisms for the nucleocapsid formation. Structure-based mutational analysis of both MARV and EBOV NPs identified key residues for helical assembly and subsequent viral RNA synthesis. Importantly, most of the residues identified were conserved in both viruses. These findings provide a structural basis for understanding the nucleocapsid formation and contribute to the development of novel antivirals against MARV and EBOV. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31420.map.gz emd_31420.map.gz | 405.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31420-v30.xml emd-31420-v30.xml emd-31420.xml emd-31420.xml | 24.7 KB 24.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_31420_fsc.xml emd_31420_fsc.xml | 18.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_31420.png emd_31420.png | 137 KB | ||
| Masks |  emd_31420_msk_1.map emd_31420_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-31420.cif.gz emd-31420.cif.gz | 6.5 KB | ||
| Others |  emd_31420_additional_1.map.gz emd_31420_additional_1.map.gz emd_31420_half_map_1.map.gz emd_31420_half_map_1.map.gz emd_31420_half_map_2.map.gz emd_31420_half_map_2.map.gz | 281.9 MB 405 MB 404.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31420 http://ftp.pdbj.org/pub/emdb/structures/EMD-31420 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31420 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31420 | HTTPS FTP |
-Validation report
| Summary document |  emd_31420_validation.pdf.gz emd_31420_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_31420_full_validation.pdf.gz emd_31420_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_31420_validation.xml.gz emd_31420_validation.xml.gz | 26.6 KB | Display | |
| Data in CIF |  emd_31420_validation.cif.gz emd_31420_validation.cif.gz | 35.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31420 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31420 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31420 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31420 | HTTPS FTP |
-Related structure data
| Related structure data |  7f1mMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10733 (Title: Marburgvirus nucleoprotein RNA complex / Data size: 3.3 TB EMPIAR-10733 (Title: Marburgvirus nucleoprotein RNA complex / Data size: 3.3 TBData #1: 2469 Falcon3 movies in TIFF [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31420.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31420.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.13 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_31420_msk_1.map emd_31420_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
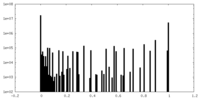
| Density Histograms |
-Additional map: #2
| File | emd_31420_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
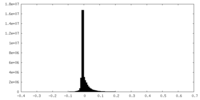
| Density Histograms |
-Half map: #2
| File | emd_31420_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
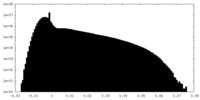
| Density Histograms |
-Half map: #1
| File | emd_31420_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Marburgvirus nucleoprotein RNA complex
| Entire | Name: Marburgvirus nucleoprotein RNA complex |
|---|---|
| Components |
|
-Supramolecule #1: Marburgvirus nucleoprotein RNA complex
| Supramolecule | Name: Marburgvirus nucleoprotein RNA complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Nucleoprotein
| Macromolecule | Name: Nucleoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Lake Victoria marburgvirus (strain Angola/2005) / Strain: Angola/2005 Lake Victoria marburgvirus (strain Angola/2005) / Strain: Angola/2005 |
| Molecular weight | Theoretical: 44.165332 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDLHSLLELG TKPTAPHVRN KKVILFDTNH QVSICNQIID AINSGIDLGD LLEGGLLTLC VEHYYNSDKD KFNTSPIAKY LRDAGYEFD VIKNADATRF LDVIPNEPHY SPLILALKTL ESTESQRGRI GLFLSFCSLF LPKLVVGDRA SIEKALRQVT V HQEQGIVT ...String: MDLHSLLELG TKPTAPHVRN KKVILFDTNH QVSICNQIID AINSGIDLGD LLEGGLLTLC VEHYYNSDKD KFNTSPIAKY LRDAGYEFD VIKNADATRF LDVIPNEPHY SPLILALKTL ESTESQRGRI GLFLSFCSLF LPKLVVGDRA SIEKALRQVT V HQEQGIVT YPNHWLTTGH MKVIFGILRS SFILKFVLIY QGVNLVTGHD AYDSIISNSV GQTRFSGLLI VKTVLEFILQ KT DSGVTLH PLVRTSKVKN EVASFKQALS NLARHGEYAP FARVLNLSGI NNLEHGLYPQ LSAIALGVAT AHGSTLAGVN VGE QYQQLR EAAYDAEVKL QRRHEHQEIQ AIAEDDEERK ILEQFHLQKT EITHSQTLAV LSQKREKLAR LAAEIENNI UniProtKB: Nucleoprotein |
-Macromolecule #2: RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3')
| Macromolecule | Name: RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') / type: rna / ID: 2 / Number of copies: 2 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 1.792037 KDa |
| Sequence | String: UUUUUU |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Concentration | 1.47 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: Apply 1.25 micro littere of sample from each side of the grid and blot for 7 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 2469 / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 2.5 µm / Calibrated defocus min: 0.5 µm / Calibrated magnification: 59000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus max: 2.25 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)