+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31358 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | PVC-Empty baseplate | |||||||||
 Map data Map data | PVC-Empty baseplate | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Photorhabdus asymbiotica (bacteria) Photorhabdus asymbiotica (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Wang X / Cheng J / Shen J / Liu L / Li N / Gao N / Jiang F / Jin Q | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Sci China Life Sci / Year: 2022 Journal: Sci China Life Sci / Year: 2022Title: Characterization of Photorhabdus Virulence Cassette as a causative agent in the emerging pathogen Photorhabdus asymbiotica. Authors: Xia Wang / Jiaxuan Cheng / Jiawei Shen / Liguo Liu / Ningning Li / Ning Gao / Feng Jiang / Qi Jin /  Abstract: The extracellular contractile injection systems (eCISs) are encoded in the genomes of a large number of bacteria and archaea. We have previously characterized the overall structure of Photorhabdus ...The extracellular contractile injection systems (eCISs) are encoded in the genomes of a large number of bacteria and archaea. We have previously characterized the overall structure of Photorhabdus Virulence Cassette (PVC), a typical member of the eCIS family. PVC resembles the contractile tail of bacteriophages and exerts its action by the contraction of outer sheath and injection of inner tube plus central spike. Nevertheless, the biological function of PVC effectors and the mechanism of effector translocation are still lacking. By combining cryo-electron microscopy and functional experiments, here we show that the PVC effectors Pdp1 (a new family of widespread dNTP pyrophosphatase effector in eCIS) and Pnf (a deamidase effector) are loaded inside the inner tube lumen in a "Peas in the Pod" mode. Moreover, we observe that Pdp1 and Pnf can be directly injected into J774A.1 murine macrophage and kill the target cells by disrupting the dNTP pools and actin cytoskeleton formation, respectively. Our results provide direct evidence of how PVC cargoes are loaded and delivered directly into mammalian macrophages. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31358.map.gz emd_31358.map.gz | 17.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31358-v30.xml emd-31358-v30.xml emd-31358.xml emd-31358.xml | 7.6 KB 7.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_31358.png emd_31358.png | 110 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31358 http://ftp.pdbj.org/pub/emdb/structures/EMD-31358 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31358 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31358 | HTTPS FTP |
-Validation report
| Summary document |  emd_31358_validation.pdf.gz emd_31358_validation.pdf.gz | 343.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_31358_full_validation.pdf.gz emd_31358_full_validation.pdf.gz | 343.4 KB | Display | |
| Data in XML |  emd_31358_validation.xml.gz emd_31358_validation.xml.gz | 6.6 KB | Display | |
| Data in CIF |  emd_31358_validation.cif.gz emd_31358_validation.cif.gz | 7.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31358 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31358 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31358 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31358 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_31358.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31358.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PVC-Empty baseplate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.356 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
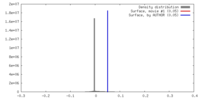
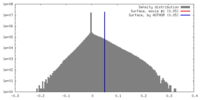
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : PVC-Empty baseplate
| Entire | Name: PVC-Empty baseplate |
|---|---|
| Components |
|
-Supramolecule #1: PVC-Empty baseplate
| Supramolecule | Name: PVC-Empty baseplate / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Photorhabdus asymbiotica (bacteria) Photorhabdus asymbiotica (bacteria) |
| Recombinant expression | Organism:  Escherichia phage EcSzw-2 (virus) Escherichia phage EcSzw-2 (virus) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 30.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 6229 |
|---|---|
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)