+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30957 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of hDisp1NNN-3C-Cleavage | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | membrane protein / LIPID TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationpatched ligand maturation / diaphragm development / embryonic pattern specification / peptide transmembrane transporter activity / dorsal/ventral pattern formation / determination of left/right symmetry / peptide transport / smoothened signaling pathway / regulation of protein secretion / protein homotrimerization ...patched ligand maturation / diaphragm development / embryonic pattern specification / peptide transmembrane transporter activity / dorsal/ventral pattern formation / determination of left/right symmetry / peptide transport / smoothened signaling pathway / regulation of protein secretion / protein homotrimerization / basolateral plasma membrane / membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.68 Å | |||||||||
 Authors Authors | Li W / Wang L | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Structural insights into proteolytic activation of the human Dispatched1 transporter for Hedgehog morphogen release. Authors: Wanqiu Li / Linlin Wang / Bradley M Wierbowski / Mo Lu / Feitong Dong / Wenchen Liu / Sisi Li / Peiyi Wang / Adrian Salic / Xin Gong /   Abstract: The membrane protein Dispatched (Disp), which belongs to the RND family of small molecule transporters, is essential for Hedgehog (Hh) signaling, by catalyzing the extracellular release of palmitate- ...The membrane protein Dispatched (Disp), which belongs to the RND family of small molecule transporters, is essential for Hedgehog (Hh) signaling, by catalyzing the extracellular release of palmitate- and cholesterol-modified Hh ligands from producing cells. Disp function requires Furin-mediated proteolytic cleavage of its extracellular domain, but how this activates Disp remains obscure. Here, we employ cryo-electron microscopy to determine atomic structures of human Disp1 (hDisp1), before and after cleavage, and in complex with lipid-modified Sonic hedgehog (Shh) ligand. These structures, together with biochemical data, reveal that proteolytic cleavage opens the extracellular domain of hDisp1, removing steric hindrance to Shh binding. Structure-guided functional experiments demonstrate the role of hDisp1-Shh interactions in ligand release. Our results clarify the mechanisms of hDisp1 activation and Shh morphogen release, and highlight how a unique proteolytic cleavage event enabled acquisition of a protein substrate by a member of a family of small molecule transporters. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30957.map.gz emd_30957.map.gz | 21.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30957-v30.xml emd-30957-v30.xml emd-30957.xml emd-30957.xml | 12.7 KB 12.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_30957.png emd_30957.png | 41.7 KB | ||
| Filedesc metadata |  emd-30957.cif.gz emd-30957.cif.gz | 6.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30957 http://ftp.pdbj.org/pub/emdb/structures/EMD-30957 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30957 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30957 | HTTPS FTP |
-Validation report
| Summary document |  emd_30957_validation.pdf.gz emd_30957_validation.pdf.gz | 521.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_30957_full_validation.pdf.gz emd_30957_full_validation.pdf.gz | 521.2 KB | Display | |
| Data in XML |  emd_30957_validation.xml.gz emd_30957_validation.xml.gz | 5.5 KB | Display | |
| Data in CIF |  emd_30957_validation.cif.gz emd_30957_validation.cif.gz | 6.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30957 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30957 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30957 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30957 | HTTPS FTP |
-Related structure data
| Related structure data |  7e2hMC  7e2gC  7e2iC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30957.map.gz / Format: CCP4 / Size: 23.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30957.map.gz / Format: CCP4 / Size: 23.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
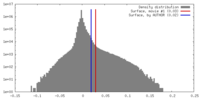
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : hDisp1NNN-3C
| Entire | Name: hDisp1NNN-3C |
|---|---|
| Components |
|
-Supramolecule #1: hDisp1NNN-3C
| Supramolecule | Name: hDisp1NNN-3C / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 150 KDa |
-Macromolecule #1: Protein dispatched homolog 1
| Macromolecule | Name: Protein dispatched homolog 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 29.497619 KDa |
| Recombinant expression | Organism: Mammalian expression vector EGFP-MCS-pcDNA3.1 (others) |
| Sequence | String: MAMSNGNNDF VVLSNSSIAT SAANPSPLTP CDGDHAAQQL TPKEATRTKV SPNGCLQLNG TVKSSFLPLD NQRMPQMLPQ CCHPCPYHH PLTSHSSHQE CHPEAGPAAP SALASCCMQP HSEYSASLCP NHSPVYQTTC CLQPSPSFCL HHPWPDHFQH Q PVQQHIAN ...String: MAMSNGNNDF VVLSNSSIAT SAANPSPLTP CDGDHAAQQL TPKEATRTKV SPNGCLQLNG TVKSSFLPLD NQRMPQMLPQ CCHPCPYHH PLTSHSSHQE CHPEAGPAAP SALASCCMQP HSEYSASLCP NHSPVYQTTC CLQPSPSFCL HHPWPDHFQH Q PVQQHIAN IRPSRPFKLP KSYAALIADW PVVVLGMCTM FIVVCALVGV LVPELPDFSD PLLGFEPRGT AIGQRLVTWN NM VKNTGYK ATLANYPFKY ADEQASSLEV LFQ UniProtKB: Protein dispatched homolog 1 |
-Macromolecule #2: Protein dispatched homolog 1
| Macromolecule | Name: Protein dispatched homolog 1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 140.427594 KDa |
| Recombinant expression | Organism: Mammalian expression vector EGFP-MCS-pcDNA3.1 (others) |
| Sequence | String: GPGSEVDWNF HKDSFFCDVP SDRYSRVVFT SSGGETLWNL PAIKSMCNVD NSRIRSHPQF GDLCQRTTAA SCCPSWTLGN YIAILNNRS SCQKIVERDV SHTLKLLRTC AKHYQNGTLG PDCWDMAARR KDQLKCTNVP RKCTKYNAVY QILHYLVDKD F MTPKTADY ...String: GPGSEVDWNF HKDSFFCDVP SDRYSRVVFT SSGGETLWNL PAIKSMCNVD NSRIRSHPQF GDLCQRTTAA SCCPSWTLGN YIAILNNRS SCQKIVERDV SHTLKLLRTC AKHYQNGTLG PDCWDMAARR KDQLKCTNVP RKCTKYNAVY QILHYLVDKD F MTPKTADY ATPALKYSML FSPTEKGESM MNIYLDNFEN WNSSDGVTTI TGIEFGIKHS LFQDYLLMDT VYPAIAIVIV LL VMCVYTK SMFITLMTMF AIISSLIVSY FLYRVVFHFE FFPFMNLTAL IILVGIGANN AFVLCDVWNY TKFDKPHAET SET VSITLQ HAALSMFVTS FTTAAAFYAN YVSNITAIRC FGVYAGTAIL VNYVLMVTWL PAVVVLHERY LLNIFTCFKK PQQQ IYDNK SCWTVACQKC HKVLFAISEA SRIFFEKVLP CIVIKFRYLW LFWFLALTVG GAYIVCINPK MKLPSLELSE FQVFR SSHP FERYDAEYKK LFMFERVHHG EELHMPITVI WGVSPEDNGN PLNPKSKGKL TLDSSFNIAS PASQAWILHF CQKLRN QTF FYQTDEQDFT SCFIETFKQW MENQDCDEPA LYPCCSHWSF PYKQEIFELC IKRAIMELER STGYHLDSKT PGPRFDI ND TIRAVVLEFQ STYLFTLAYE KMHQFYKEVD SWISSELSSA PEGLSNGWFV SNLEFYDLQD SLSDGTLIAM GLSVAVAF S VMLLTTWNII ISLYAIISIA GTIFVTVGSL VLLGWELNVL ESVTISVAVG LSVNFAVHYG VAYRLAPDPD REGKVIFSL SRVGSAMAMA ALTTFVAGAM MMPSTVLAYT QLGTFMMLIM CISWAFATFF FQCMCRCLGP QGTCGQIPLP KKLQCSAFSH ALSTSPSDK GQSKTHTINA YHLDPRGPKS ELEHEFYELE PLASHSCTAP EKTTYEETHI CSEFFNSQAK NLGMPVHAAY N SELSKSTE SDAGSALLQP PLEQHTVCHF FSLNQRCSCP DAYKHLNYGP HSCQQMGDCL CHQCSPTTSS FVQIQNGVAP LK ATHQAVE GFVHPITHIH HCPCLQGRVK PAGMQNSLPR NFFLHPVQHI QAQEKIGKTN VHSLQRSIEE HLPKMAEPSS FVC RSTGSL LKTCCDPENK QRELCKNRDV SNLESSGGTE NKAGGKVELS LSQTDASVNS EHFNQNEPKV LFNHLMGEAG CRSC PNNSQ SCGRIVRVKC NSVDCQMPNM EANVPAVLTH SELSGESLLI KTL UniProtKB: Protein dispatched homolog 1 |
-Macromolecule #3: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 3 / Number of copies: 8 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 4 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 7.5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.68 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 63043 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)