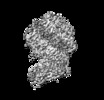
Entry Database : EMDB / ID : EMD-30816Title Cryo-EM structure of SARS-CoV2 RBD-ACE2 complex Complex : Cryo-EM structure of SARS-CoV2 RBD-ACE2 complexComplex : ACE2Protein or peptide : Angiotensin-converting enzyme 2Complex : SARS-CoV2 RBDProtein or peptide : Spike glycoproteinLigand : ZINC IONLigand : CHLORIDE IONLigand : 2-acetamido-2-deoxy-beta-D-glucopyranoseLigand : water / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human) / Method / / Resolution : 2.8 Å Wang J / Lan J Journal : Biophys Rep / Year : 2022Title : Reduced graphene oxide membrane as supporting film for high-resolution cryo-EMAuthors : Liu N / Zheng LM / Xu J / Wang J / Hu CX / Lan J / Zhang X / Zhang JC / Xu K / Cheng H / Yang Z / Gao X / Wang XQ / Peng HL / Chen YN / Wang HW History Deposition Dec 22, 2020 - Header (metadata) release Dec 29, 2021 - Map release Dec 29, 2021 - Update Oct 23, 2024 - Current status Oct 23, 2024 Processing site : PDBj / Status : Released
Show all Show less
 Open data
Open data Basic information
Basic information Map data
Map data Sample
Sample Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human) /
Homo sapiens (human) / 
 Authors
Authors Citation
Citation Journal: Biophys Rep / Year: 2022
Journal: Biophys Rep / Year: 2022 Structure visualization
Structure visualization Movie viewer
Movie viewer SurfView
SurfView Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links emd_30816.map.gz
emd_30816.map.gz EMDB map data format
EMDB map data format emd-30816-v30.xml
emd-30816-v30.xml emd-30816.xml
emd-30816.xml EMDB header
EMDB header emd_30816.png
emd_30816.png emd-30816.cif.gz
emd-30816.cif.gz http://ftp.pdbj.org/pub/emdb/structures/EMD-30816
http://ftp.pdbj.org/pub/emdb/structures/EMD-30816 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30816
ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30816 emd_30816_validation.pdf.gz
emd_30816_validation.pdf.gz EMDB validaton report
EMDB validaton report emd_30816_full_validation.pdf.gz
emd_30816_full_validation.pdf.gz emd_30816_validation.xml.gz
emd_30816_validation.xml.gz emd_30816_validation.cif.gz
emd_30816_validation.cif.gz https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30816
https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30816 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30816
ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30816
 Links
Links EMDB (EBI/PDBe) /
EMDB (EBI/PDBe) /  EMDataResource
EMDataResource Map
Map Download / File: emd_30816.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
Download / File: emd_30816.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Sample components
Sample components Homo sapiens (human)
Homo sapiens (human)
 Homo sapiens (human)
Homo sapiens (human) Homo sapiens (human)
Homo sapiens (human)
 Homo sapiens (human)
Homo sapiens (human)

 Processing
Processing Sample preparation
Sample preparation Electron microscopy
Electron microscopy FIELD EMISSION GUN
FIELD EMISSION GUN
 Image processing
Image processing Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)